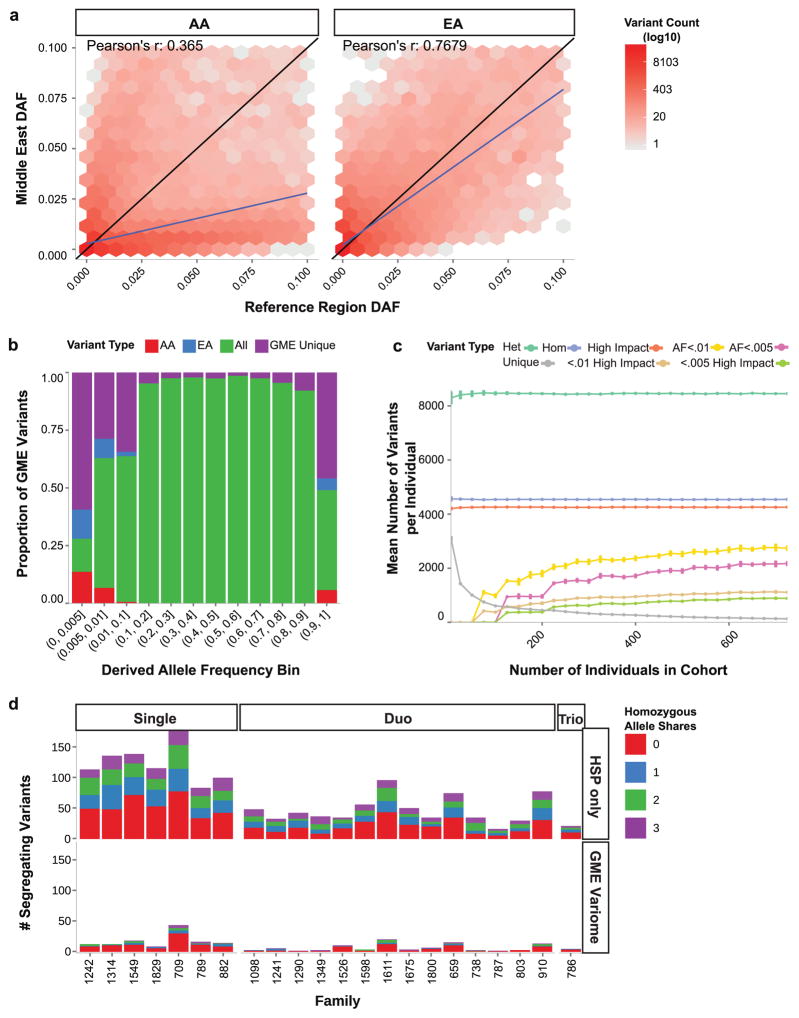
Figure 4. GME Variome facilitates the discovery of Mendelian disease genes.
a–b. Comparison of rare derived allele frequencies (DAF) between GME and Exome Sequencing Project (ESP). AA: African American, EA: European-American. Hexagonal bins shaded by log number of variants within each bin. Pearson’s r suggests GME DAFs were not accurately estimated by AA or EA populations.
b. The majority of variants in the rarest DAF bins were unique to the GME. AA: found only in GME and AA. EA: found only in GME and EA. All: found in GME, EA and AA. GME Unique: found only in GME.
c. Change in per-individual burden of eight variant classes as a function of increasing the number of individuals incorporated into the GME Variome cohort. As sample size increased there was a drop in the number of unique variants, along with more accurate estimation of DAFs for rare variants. Bootstraps were sampled with replacement for 100 iterations to calculate standard errors. “High impact”: variants meeting predicted deleteriousness thresholds (see Methods).
d. Number of candidate variants for 20 families, meeting segregation and deleteriousness filtering criteria, using DAFs derived from Hereditary Spastic Paraplegia (HSP)-only families (top) or also incorporating the GME Variome (bottom). Single, Duo, Trio: families with one, two or three affected members. Colors: number of individuals sharing the variant. “0”: no other individuals carried the allele, etc. Analysis was performed using this threshold for the number of individuals sharing alleles (0,1,2,3). Note drop in number of segregating variants for any given family after the GME Variome was applied.