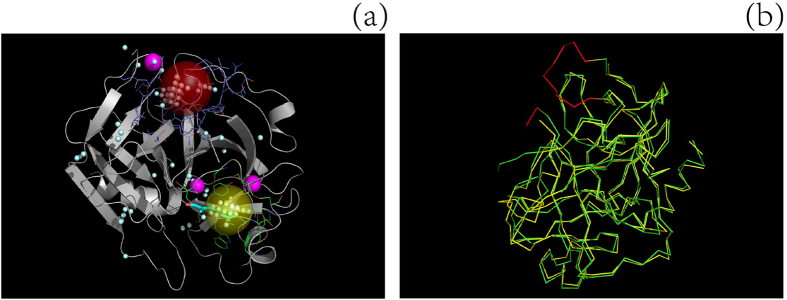
Figure 4.
Comparison of the ligand-binding sites on a protein ((a) ID: 1CHG) predicted using different methods and alignment of the ligand-unbound structure with the ligand-bound structure to obtain missing Cα backbone atoms (b). The actual ligand-binding site (yellow ball) was the second cavity after re-ranking by the MF-PLB, whereas the first cavity (red ball) assigned by the MF-PLB is 16.63 Å away from the ligand (OAC). After examining the protein pdb file, we found that some residues were missing (red part in b) and that the presence of these missing residues would result in an abnormal cavity (red ball in a). We added these missing residues to protein 1CHG by aligning 1CHG to its ligand-bound form (3GCH) using PyMOL. The red-ball binding site was not obtained after the pocket grids were calculated by Ligsite-cs, and the first cavity assigned by the MF-PLB after the addition of the missing residues was the actual ligand-binding site. The MF-PLB values for the red-ball cavity and the yellow-ball cavity are 23.93 and 20.55, respectively. All pocket grids calculated by Ligsite-cs are shown as cyan grids.