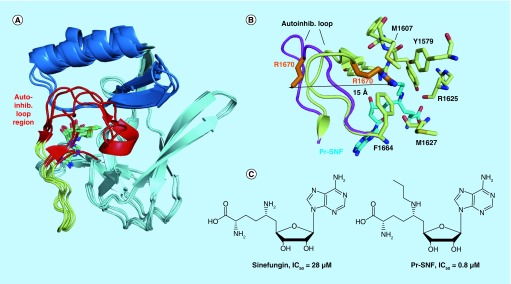
Figure 3. . Structures of H3K36-specific S-adenosyl methionine domains and SAM-competitive inhibitors.
(A) Overlay of the core SET (pale cyan), SET-I (sky blue) and post-SET (pale yellow) subdomains of NSD1 (PDB code 3OOI), NSD3 (4YZ8), ASH1L (4YNM) and SETD2 (4H12). The structurally variable autoinhibitory loop region is colored in red. (B) Binding of the N-propyl sinefungin (Pr-SNF) inhibitor to SETD2 causes opening of the autoinhibitory loop. Autoinhibitory loop conformation is shown with S-adenosyl homocysteine bound (pale yellow, 4H12) and with Pr-SNF bound (magenta, 4FMU). Steric clash between the propyl moiety of Pr-SNF (cyan) and the Arg1670 sidechain (orange) causes Arg1670 to flip out a distance of 15 Å. Residues stabilizing Arg1670 in the putative substrate lysine-binding channel in the S-adenosyl homocysteine bound form of SETD2 are shown in pale yellow sticks. (C) Chemical structures of SETD2 inhibitors sinefungin and Pr-SNF. In vitro IC50 values for SETD2 are listed.