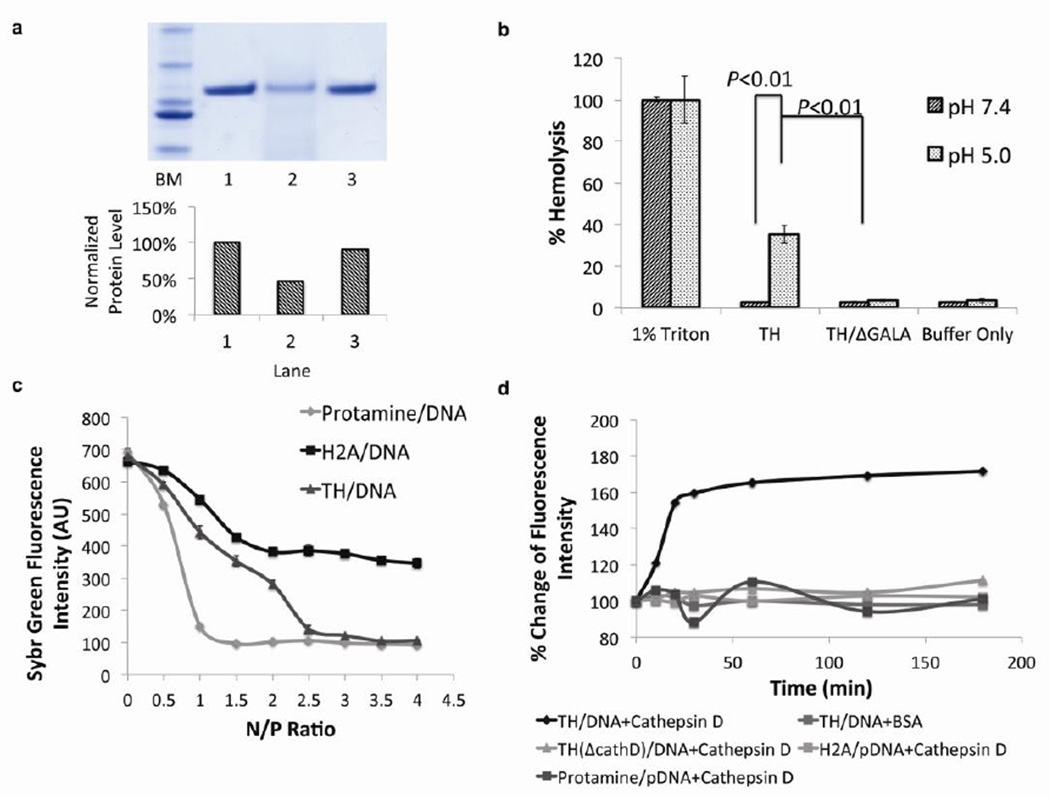
Figure 2. The incorporation of multiple functional amino acid sequences in one molecule has preserved the functionality of each sequence.
a) SDS-PAGE of digested TH with cathepsin D protease. Lane 1: ladder marker, Lane 2: undigested TH, lane 3: TH digested by cathepsin D. Lane 4: TH/ΔCathD digested by cathepsin D. Bottom chart: Quantification of relative amount of protein based on densitometry analysis. The incorporation of cathepsin D substrate in the TH facilitated the degradation in the presence of corresponding enzyme. b) Hemolysis activity of TH at acidic (pH=5.0) or neutral (pH=7.4) pH. 1% TritonX-100 was used as a positive control (representing 100% hemolysis) and buffer alone was used as a negative control (n=3). The endosome disruptive sequence, GALA, in the TH induced lysis of the membrane exclusively at an acidic pH. Error bars show mean ± s.d. Student t-test. c) SYBR Green exclusion assay. Plasmid DNA with SYBR Green I stain was complexed with protamine, H2A peptide (37mer) or TH at various N/P ratios. (n=3). TH-mediated DNA condensation was equivalent to that of protamine, whereas H2A could only condense DNA into a less compact degree. Error bars show mean ± s.d. d) Cathepsin D induced decondensation. Plasmid DNA was complexed with TH, TH/ΔCathD, H2A peptide and protamine at optimal N/P ratio, followed by cathepsin D incubation. The incorporation of cathepsin cleavage enabled the TH to respond to the stimulus in a prompt way, facilitating the release of nucleic acids in an enzyme dependent manner.