Abstract
Alterations in the composition of the commensal microbiota have been observed in many complex diseases. Understanding the basis for these changes, how they relate to disease risk or activity, and the mechanisms by which the symbiotic state of colonization resistance and host homeostasis is restored is critical for future therapies aimed at manipulating the microbiota.
INTRODUCTION
Normal mammalian structure and function is significantly dependent on commensal microbes, which normally colonize many sites of the body but especially in the gastrointestinal tract. The development of these indigenous commensal microbial communities primarily begins in the first moments of our lives and depends on the inoculum with founder species and, ultimately, diversification of bacterial communities brought about by encounters with new microbial species as well as environmental and host influences (1, 2). In the end, this commensal ontogeny results in an individual microbial fingerprint (3). In adults, the composition of the intestinal microbiota is relatively stable but can undergo dynamic changes as a result of its interactions with diet, genotype/epigenetic composition, and immune-metabolic function (4). The consequences of these various interactions can vary markedly. For example, the ingestion of inulin-containing prebiotics, ingested substances that influence microbial function and consequently community composition, promotes the proliferation of short-chain fatty acid (SCFA)–producing bacteria with immune and nonimmune consequences (4). Other effects are far more reaching. These include an infection with an enteropathogen or repetitive exposures to broad-spectrum antibiotics, both of which can change profoundly the composition of the microbiota in a manner that may be detrimental to the host, leading to so-called dysbiosis. Such environmentally induced alterations in the composition of the commensal microbiota may moreover be irreversible (5–7), making it possible that they are associated with functional changes in the host, which predispose to the development of chronic disease. It is interesting in this regard that it remains a conundrum why diseases often arise in an age- and site-specific manner [for example, right side of the colon in inflammatory bowel disease (IBD) in the second decade of life] (8) or why distinct complex diseases often involve common immune pathologic pathways (9). These observations raise the question of whether mucosal dysbiosis induced by host and/or environmental factors can play a role in local or systemic disease development. Providing answers to these questions is important because it may help enable the ability to predict disease development or support the rationale for restoration of the microbiota to a zone of normality to reestablish health. In this Review, we will consider these questions with a specific focus on the gastrointestinal tract where most information is available.
“COLONIZATION RESISTANCE” AND THE RISK OF ENTEROPATHOGENIC INFECTIONS
Once established, the indigenous microbiota provides many crucial functions to the host, consistent with the coevolved symbiotic relationship between microbes and humans. These have been reviewed elsewhere (10–12) and include the contribution to digestion (such as the ability of microbes to break down host nondigestible polysaccharides) and its secondary benefits (the generation of SCFA), the metabolism of xenobiotics, which aids in protection from environmental toxins, and, as briefly discussed here, colonization resistance. The origin of the concept of colonization resistance dates to the studies of Dubos in 1965 who demonstrated the role of the indigenous microbiota in antagonizing colonization with a potential pathogen (13). It is now appreciated that this protective function is probably woven into the many symbiotic functions of the commensal microbiota (14). These include (i) the competitive metabolic challenge faced by a microbial invader and (ii) the role of the commensal microbiota in stimulating the maintenance of a protective mucosal barrier that includes many structural (for example, tight junctions) and functional (for example, neutrophil transmigration, antimicrobial peptides, and mucus) components (15). Colonization resistance provides broad protection from bacteria, viruses, and possibly other classes of pathogens (16). True pathogens are equipped to overwhelm and evade this barrier by many different mechanisms such as the ability to invade the epithelium, subvert the immune response, and/or outcompete the commensal microbiota for essential nutritional substrates (17). A potentially illustrative strategy is the ability of the pathogen to induce a profound inflammatory state that is associated with high-level production of antimicrobial peptides to which the pathogen may be resistant (18). Although not directly demonstrated, this likely results in a disintegration of the microbial communities that provide natural defense, access of the invader to a nutrient-rich environment enriched by the inflammatory process itself, and the outgrowth of potential inflammatory allies such as Proteobacteria, which also survive and thrive in such a milieu (14).
In many ways, antibiotic treatment replicates infection with a pathogen by reducing targeted classes of microbiota. As observed in mice and in antibiotic-treated patients undergoing hematopoietic stem cell transplant (19), this allows resistant symbiotic commensalists to emerge with pathogenic properties, so-called pathobionts (20). Toxin-producing bacteria such as Clostridium difficile may also find an advantage when colonization resistance is reduced (21). Although antibiotics are characteristically administered as therapy to drive out C. difficile or other toxin-producing pathogens such as Campylobacter jejuni and Salmonella typhimurium, emerging evidence suggests that strategies that reestablish commensalism (for example, restraint from antibiotics or even bacterial therapy through fecal transplantation) may be just as effective (22) This is not to say that antibiotics are not potentially critical to the treatment of antibiotic-associated C. difficile or other infections, but that colonization resistance imposed by the microbiota is an important mediator of disease resistance.
Mechanisms unrelated to antibiotics are also likely to contribute to dysbiosis induced by the loss of colonization resistance because C. difficile overgrowth and its clinical consequences can occur in the absence of any known antibiotic exposure. Indeed, enteropathogenic infection per se, inflammation itself, and potentially malnutrition can cause disruptions of commensal microbiota, both in animal models and probably in humans as discussed below (23, 24). Erosion of colonization resistance and increased susceptibility to the effects of true pathogens and pathobionts is therefore just one of the many consequences that flow secondarily from microbial dysbiosis.
Clearly, not every person who is exposed to antibiotics and experiences privation of beneficial classes of microbiota, or who carries true pathogens, necessarily develops disease (25). This is illustrated by young children infected with C. difficile. Thirty to up to 90% of healthy infants are colonized with C. difficile during the first year of life and yet puzzlingly show no disease symptoms (26). Moreover, many individuals who develop a symptomatic infectious disease are able to evict the pathogen (or revert the pathobiont), restore tissue structure and function, and presumably reconstitute symbiotic commensal microbial communities. There is thus a broad range of disease tolerance among individuals (27). Elucidating the factors that determine such disease tolerance or susceptibility in instances of clear-cut commensal breakdown as well as the mechanisms used by the host to reestablish a normal state of microbial self is of great importance. Such insights will serve to help develop future preventative or therapeutic strategies not only for infectious diseases but also for chronic diseases that are due to or facilitated by states of dysbiosis (Fig. 1).
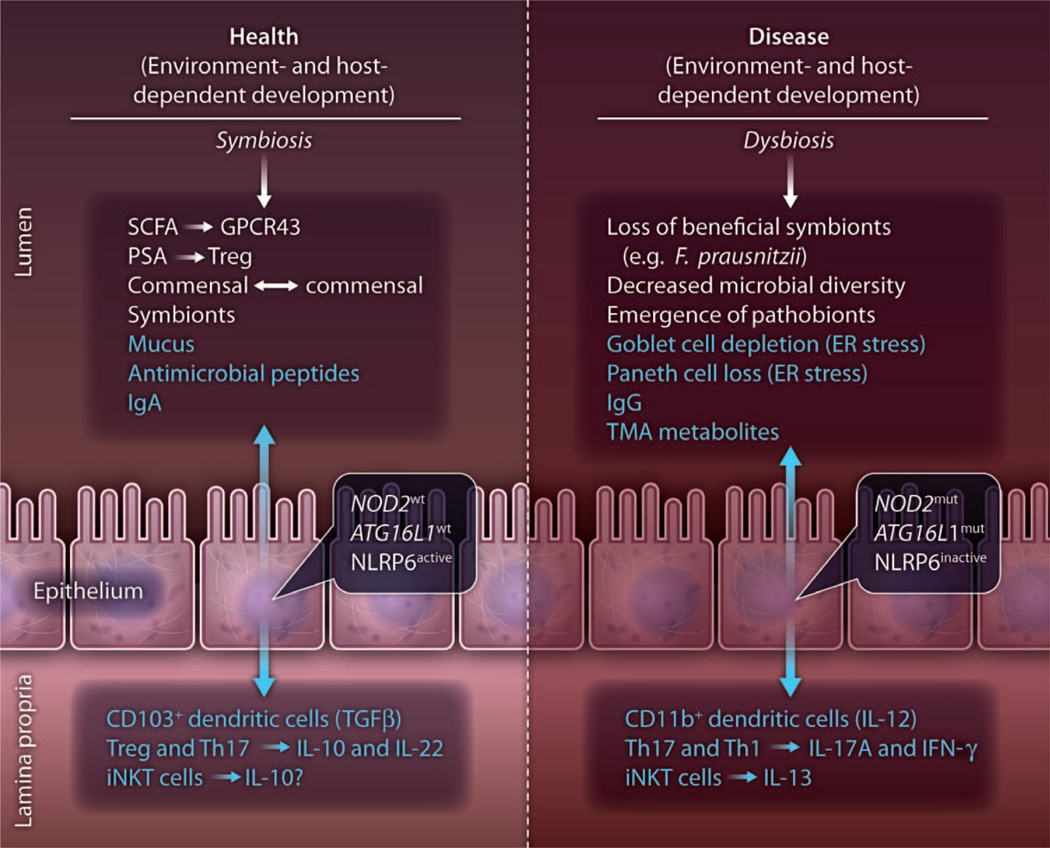
Fig. 1.
Factors at the commensal-host interface associated with health and disease. Environmental and host factors (which are under genetic control) determine the composition and functional consequences of the components within the lumenal milieu. The lumenal milieu consists of commensal microbiota and their products and secreted factors of the host (blue). These lumenal states are metastable at any given point in time and likely age-dependent. Health is associated with symbiosis of the commensal microbiota and host responses (lumenal, epithelial, and subepithelial), which are a reflection of homeostasis. Disease on the other hand is characterized by dysbiosis of the commensal microbiota and the corollary host responses. Whether symbiosis or dysbiosis is a primary or secondary factor in disease development remains an open question. The ultimate development of disease at any given set point in this model is dependent on many factors, which determine tissue tolerance and include genetic susceptibility among others. GPCR43, G protein–coupled receptor 43; PSA, polysaccharide antigen A; Treg, T regulatory cell; ER, endoplasmic reticulum; wt, wild-type allele; mut, mutant allele; TMA, trimethylamine; Th, T helper; IL, interleukin; TGFβ, transforming growth factor–β; IFNγ, interferon-γ; iNKT, invariant natural killer T.
DYSBIOSIS AND COMPLEX DISEASE: CAUSE OR CONSEQUENCE
Our intestinal microbiota is highly individual, tuned by our genetic makeup and environment. In health, our microbial residents contribute to the physiology of a number of intestinal and extraintestinal body systems. Not surprisingly, changes in the composition of the intestinal microbiota have been linked to diverse, complex diseases including IBD (encompassing Crohn’s disease and ulcerative colitis), metabolic disease, type 1 diabetes mellitus, allergy, asthma, and neurologic and cardiovascular disease (28, 29). Some have hypothesized that environmental “flatteners” derived from the forces of globalization, such as extensive antibiotic use, changes in the food supply, and the increased intermixing of humans across international borders, may result in major ecological disruptions in the commensal microbiota that contribute to the development of such diseases (30). Despite the growing number of reports linking alterations in the microbiota to inflammatory disease, it remains to be established whether these are primary or secondary events. This is complicated by the fact that very large studies are required to distinguish disease-associated change from the profound amount of interindividual variation observed in the microbiome. For example, monozygotic twins share only 40% of phylotypes and exhibit even more divergence when characterized at the level of the metagenome and metatranscriptome (31).
INFLAMMATORY BOWEL DISEASE
There is abundant evidence that the microbiota plays a key role in the pathogenesis of Crohn’s disease and ulcerative colitis, both chronic relapsing diseases of the gastrointestinal tract. In mouse models, the microbiota is often required for the development of colitis in genetically susceptible hosts (32). However, the microbiota can also provide protection against the development of colitis. For example, germ-reduced mice generated by antibiotic administration or germ-free mice that are raised under sterile conditions exhibit more severe disease in a dextran sodium sulfate (DSS)–induced colitis model that is dependent on the innate immune system (33, 34). This increased susceptibility is the result of decreased production of SCFA in the absence of the commensal microbiota. SCFA engagement of the G protein (heterotrimeric guanine nucleotide–binding protein)–coupled receptor 43 (GPCR43) on polymorphonuclear leukocytes acts to diminish their infiltration into tissues and, consequently, protects against inflammation (34). Enhanced susceptibility of germ-free or germ-reduced mice to colitis models dependent on the adaptive immune system has also been observed. Invariant natural killer T (iNKT) cells, which recognize lipid antigens in the context of CD1d on antigen-presenting cells, are increased in the colonic tissues of germ-free mice and cause profound inflammation when triggered by stimulating environmental factors in oxazolone-induced colitis (35). Thus, both the innate and the adaptive limbs of the mucosal immune response are stimulated or restrained by the effects of intestinal microbes. Although more difficult to ascribe a direct link, some patients with Crohn’s disease similarly exhibit decreased inflammation in response to antibiotic therapy (36). Similarly, diversion of the fecal stream in IBD patients ameliorates intestinal inflammation, which reappears in a significant proportion of patients after restoration of the commensal microflora (36). Hence, IBD may be viewed as a maladaptation of host commensal mutualism in which genetic and environmental factors conspire to promote the accumulation of microbial communities capable of driving disease as well as loss of protective groups of microorganisms, with the end result being chronic intestinal inflammation.
Support for this comes from a recent large observational study showing that frequent antibiotic use during the first year of life increased the risk of developing Crohn’s disease (37). Although not necessarily causative, these results suggest that early-life host-microbiota interactions are especially important in setting the threshold of intestinal homeostasis (38, 39). Similarly, later-life emergence of pathobionts such as adherent-invasive Escherichia coli, as discussed below, or loss of protective microorganisms such as Faecalibacterium prausnitzii, a Firmicute whose absence is linked to postoperative relapse of Crohn’s disease, may be important determinants of disease activity (40, 41). Thus, changes in the microbiota may occur before or after development of inflammation and affect susceptibility to and the phenotypic manifestations of disease (Fig. 2).
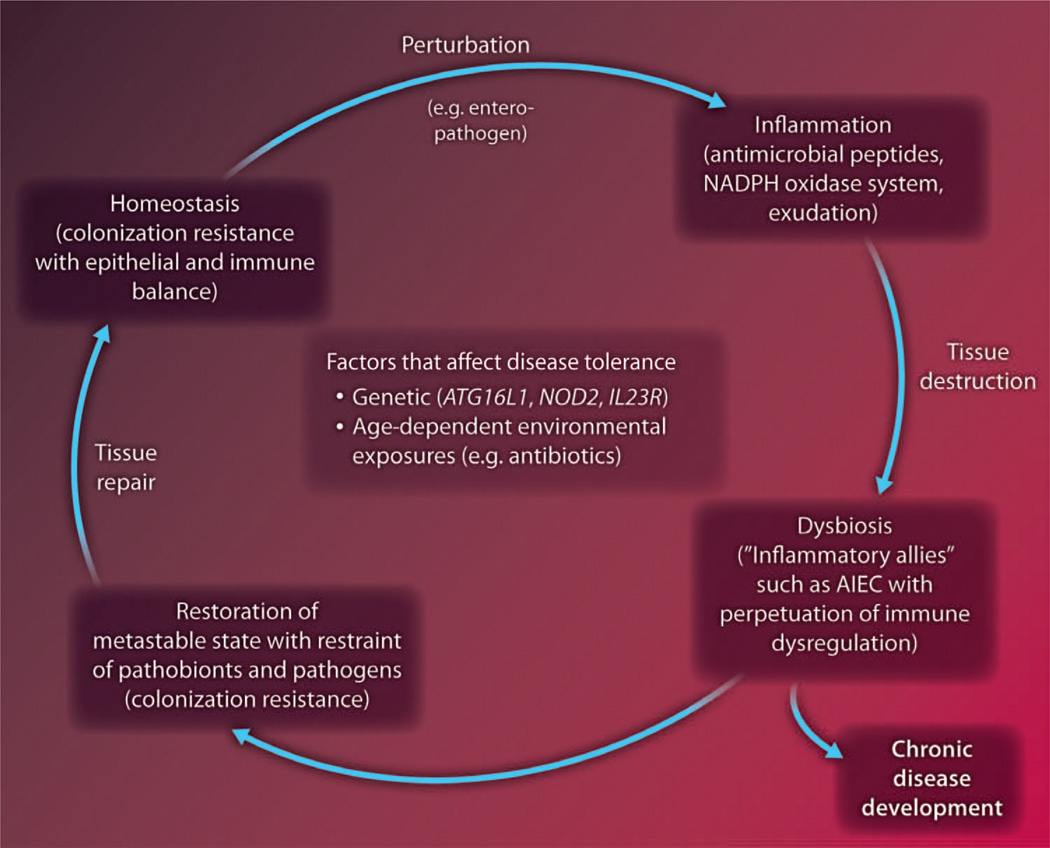
Fig. 2.
Cycles of equilibrium and imbalance at the commensal-host interface. Homeostasis is associated with symbiotic commensalism and colonization resistance together with epithelial and immune balance. Perturbations, such as enteropathogenic invasions, disrupt this balance by inducing inflammation, resulting not only in tissue destruction but also breakdown in the state of commensalism and, consequently, dysbiosis. Dysbiosis can be associated with the evolution of pathobionts that function as inflammatory allies (for example, Proteobacteria), which are able to flourish in the inflammatory milieu and further promote inflammation induced by the invading pathogen. Some types of perturbations, such as antibiotics, can lead directly to breakdown in commensalism and subsequent colonization by pathobionts and pathogens that are able to take advantage of the niche (for example, C. difficile). Under normal circumstances, the host can restore a metastable state of commensalism associated with control of pathobionts and pathogens, reestablishment of colonization resistance, and tissue repair. Individuals who are unable to accomplish this develop chronic disease. Factors that affect each of these hypothetical parts of this health-disease cycle include genetics, age, and environmental experiences among others. NADPH, reduced form of nicotinamide adenine dinucleotide phosphate; AIEC, adherent-invasive E. coli.
Results from mouse models and genome-wide association studies have converged to identify key pathways involved in intestinal homeostasis that are altered in IBD. About 100 distinct susceptibility loci have been identified in IBD, and these map to diverse pathways including intestinal barrier function, host defense, and immune regulation (42, 43). In Crohn’s disease, there is increasing evidence for dysfunction of Paneth cells, specialized intestinal epithelial cells located at the base of the crypts within the small intestine, leading to reduced antimicrobial peptide production and increased local proinflammatory activity. Cells expressing variants of the cytosolic pattern recognition receptor NOD2 (CARD15) that are associated with an increased risk of Crohn’s disease exhibit reduced activation of nuclear factor κB in response to intestinal bacteria and impaired secretion of antimicrobial peptides called α-defensins by Paneth cells (44). This alteration in antimicrobial immunity in the intestine of Crohn’s disease patients may account for the observation that these individuals also display dysbiosis characterized by increased Bacteroidetes and Firmicutes in the ileum (45). Such observations reinforce the notion that Paneth cell function plays an important and perhaps essential role in regulating access of microbes to the epithelial cell surface (46). Paneth cell abnormalities are observed in patients or mice expressing the risk allele of the autophagy gene ATG16L1, a multifunctional gene that controls defense against intracellular infection, secretory pathways, and the activity of inflammasomes within the cell (47). In mice, these defects depend on the presence of specific types of norovirus infection (48). It remains to be determined whether these effects of norovirus infection in a susceptible host are through its ability to affect the host immune response, epithelial cell function, and/or the composition of the microbiota. Nevertheless, these studies support the primary importance of a microbial factor in the development of IBD within a specific genetic context.
These and other IBD-related deficits in host defense may allow both a breakdown in the compartmentalization of the intestinal microbiota with increases in intestinal epithelial cell–associated bacteria and bacterial translocation into the intestinal mucosa. IBD is associated with a decrease in the complexity of the commensal microbiota similar to that observed during enteropathogenic infections and chemically induced colitis (24, 49, 50). This suggests that the inflammation seen in IBD is associated with a loss in colonization resistance. Indeed, 16S ribosomal RNA sequencing has revealed not only alterations in the diversity of the microbiota in IBD, with reductions in the two major phyla Bacteroidetes and Firmicutes, but also increases in adherent bacteria, particularly Enterobacteriaceae and other Proteobacteria such as adherent-invasive E. coli (24, 49, 51, 52). What may be important is not the presence of one particular bacteria but the ratio of pathogenic to more protective species. This is consistent with results showing that monozygotic twins with Crohn’s disease exhibit an increased ratio of adherent-invasive E. coli to F. prausnitzii, the latter of which has anti-inflammatory functions (53). Similarly, analysis of the mucosal microbiota in patients with ulcerative colitis or their unaffected siblings demonstrated dysbiosis, including loss of microbial richness, which correlated with a reduction in host responses that are important for barrier protection (52). Although provocative, the challenge will be to distinguish whether these changes are primary events involved in the etiology of disease or a correlate of inflammation. Indeed, Proteobacteria differentially accumulate in diverse inflammatory settings including colitis, asthma (54), and chronic gastritis (55). This suggests that such classes of bacteria may be specifically adapted to exploit the inflammatory environment at the expense of other commensal bacteria.
Resolution of the cause or consequence debate requires the use of mouse or other relevant animal models to directly test the causality of changes in the microbiota and intestinal inflammation. A number of recent studies show that specific genetic defects that predispose to increased susceptibility to colitis results in shifts in the microbiota that are in some cases transmissible. Thus, mice deficient in the innate immune system sensor, the NLRP6 inflammasome, have dysbiosis of the microbiota with increased representation of bacterial phyla such as Bacteroidetes (Prevotellaceae) and TM7 (56). These mice are more susceptible to chemically induced colitis, a phenotype that could be transferred to adult wild-type mice by the microbiota (56). Similarly, mice deficient in T cells, B cells, and the transcription factor T-bet exhibit an altered microbiota and develop an ulcerative colitis–like disease that is transmissible through the microbiota (57). Transfer of a mild form of the disease to wild-type mice correlates with the presence of Proteus mirabilis and Klebsiella pneumoniae but also requires additional and as yet unidentified components of the commensal microbiota. Along these lines, a recent study in mice has shown that colitis-inducing species are not necessarily those with the largest changes in colonization levels but rather are members of the indigenous microbiota that acquire pathogenic features only in specific host genetic and environmental contexts, that is, pathobionts (58). Together, data from mouse models suggest that there may be no universal colitogenic bacteria but that distinct pathobionts emerge from a dysbiotic commensal microbiota that are driven by a particular genetically determined, environmentally induced maladapted host response. This individuality poses challenges for treatment strategies aimed at targeting colitogenic bacteria and suggests that identification of pathobionts in IBD will require analysis of genetically defined patient subgroups displaying early clinical signs.
ALLERGY AND ASTHMA
The incidence of allergic disease and asthma is rising to epidemic proportions in the developed world. The realization of the impact of environmental factors such as the size of households, the number of older siblings in the family, the global decrease of infectious diseases, and more recently, exposure to livestock on the development of allergy and asthma has led to the hygiene hypothesis (59–61). This is now interpreted as the effect of Western life-styles: Increased sterility leads to reductions in perinatal microbial exposure, which impairs microbiota-driven protective mechanisms such as immune tolerance. Although most attention has focused on reduced exposure to exogenous microbes, there is also evidence of missing indigenous microbiota because of reductions in mother-to-child transmission. One particularly noteworthy organism is Helicobacter pylori, a member of our ancestral microbiota that dominates the gastric niche. Once present in most human individuals, antibiotic use and reduced transmission have led to a rapid decline in the frequency of individuals who harbor H. pylori, estimated to be below 10% in Western-born children (5). Although a causative agent in gastric ulcer and cancer in some individuals, H. pylori colonization in early life has been linked to enhanced gastric physiology and immunity. It has been proposed that the recent rapid decline in H. pylori colonization has contributed to the similarly rapid increases in allergic and metabolic disease (5). Indeed, epidemiological evidence shows that allergic disease and asthma are inversely correlated with the presence of H. pylori, and experimental infection with H. pylori protects against allergic asthma in a mouse model (62). Of course, H. pylori is one member of the indigenous microbiota that we can measure, and it remains to be established whether other poorly characterized or as yet unidentified microbial components may also be protective against the development of these diseases.
Despite impressive epidemiological evidence, little is known about the mechanisms by which gastrointestinal microbiota prevent local inflammation or allergic disease at distant sites such as the lungs and skin that are affected in allergic and asthmatic disease. Evidence from mouse models suggests that metabolic activity or constituents of particular microbes may protect against disease. For example, polysaccharide antigen A of Bacteroides fragilis (63) or SCFA (34) may prevent immune pathology in the intestines, lungs, joints, and central nervous system by promoting immune-suppressive T regulatory cells or controlling neutrophil function, respectively. Recent studies, however, reveal a distinct mechanism that directly supports the hygiene hypothesis. In mice, neonatal but not late-life exposure to commensal microbiota is necessary to prevent epigenetic changes that induce expression of the Cxcl16 gene, which encodes a chemokine that binds CXCR6 in the colonic and lung epithelium (35). In the absence of this microbial-induced restraint on CXCL16 expression, CXCR6+ iNKT cells accumulate in the colon and lung where later-life exposure to environmental factors leads to the induction of colitis and asthma, respectively. Consistent with this, early, but not late, antibiotic exposure affects later-life susceptibility of mice to allergic asthma (64). In another model system, the microbiota were shown to protect from allergic disease through reductions in immunoglobulin E (IgE)–mediated mobilization of basophil precursors (65). Understanding the commensal microbial organisms within the intestines and potentially lungs that are responsible for these protective immune changes may provide new routes to prevent or inhibit inflammation at barrier surfaces and restore homeostasis. Moreover, they will inform the age of the host at which such interventions can be anticipated to be successful (35).
METABOLIC DISEASE: DIABETES, OBESITY, AND CARDIOVASCULAR DISEASE
Increases in metabolic diseases such as obesity and insulin resistance worldwide have traditionally been explained by the host’s intake of higher calorific Western-style diets. However, focus has also shifted to include the impact of the microbiota on our metabolic health [see Review by Holmes et al. (66)]. This is because the intestinal microbiota through its own metabolic activity not only can provide a significant quantity of the host’s caloric content but also can be a major contributor of biologically active metabolites that can be detected both locally in the colon and in systemic sites such as the urine [see Review by Nicholson et al. (67)].
Evidence in favor of a causal link between the microbiota and obesity comes primarily from mouse models. For example, germ-free mice are resistant to high-fat diet–induced obesity because of a lack of fermenting bacteria that can process complex polysaccharides and resultant reduced SCFA production. Colonization of germ-free mice with fermenting bacteria such as Bacteroides thetaiotaomicron together with organisms that promote the fermentation process such as Methanobacterium smithii restored weight increase and obesity that was dependent on the SCFA receptor, GPCR41 (68). Similarly, changes in the microbiota and bacterial metabolic pathways occur in mice with genetic lesions in signaling pathways that regulate leptin, a hormone critical for the regulation of appetite and metabolism (67). These changes in the microbiota are associated with uncontrolled food intake and obesity. Markedly, transfer of the microbiota from leptin-deficient mice to wild-type mice led to a milder form of adiposity, which indicates that the microbiome itself can be a primary driver of metabolic disease. Another causative example of the microbiota driving metabolic disease is mice lacking the microbial sensor Toll-like receptor 5 (TLR5). These mice develop metabolic disease with hyperlipidemia, hypertension, insulin resistance, and weight gain. Tlr5−/− mice harbored an altered intestinal microbiota, which could transfer the metabolic syndrome to wild-type mice (69). Why TLR5 deficiency causes an obesity-associated microbiome is currently unknown.
The influence of the microbiota on metabolism, however, goes both ways, at least in mice. Mice fed a high-fat diet have altered bacterial community structure, which leads to reduced diversity and a phylogenetic shift from Bacteroidetes to Firmicutes. This change is associated with the presence of increased fermenters in the gut and an increased representation of bacterial pathways associated with polysaccharide fermentation and SCFA production. This functional adaptation has been coined an “obesity-associated microbiome” because of its increased capacity for energy harvest (70).
Some mechanistic insight into immune-mediated control of metabolism comes from the finding that B cells and IgA, a molecule important for the appropriate compartmentalization of the microbiota, influence the balance of immune and metabolic pathways in intestinal epithelial cells through a microbiota-dependent mechanism (71). In the presence of IgA, the intestinal microbiota promotes the expression of genes involved in lipid metabolism and storage. In the absence of IgA, however, there is a shift toward the expression of genes involved in host defense and inflammation. Although not directly demonstrated, deficiency in IgA may thus lead to a redistribution of microbial communities toward enhanced proportions of mucosa-associated bacteria. The resulting compensatory adaptation of the intestinal epithelial cell response may then veer toward host defense and away from a homeostatic metabolic response. Regardless of the mechanism, these data illustrate the connectivity between immunity, the microbiota, and metabolism, an area that is only starting to be unraveled.
Despite convincing data in mouse models, the link between the microbiota and metabolic disease in humans is less compelling. There are reports of alterations in bacterial phyla in lean versus obese individuals, but other studies have not found this (72). Of course, human studies are confounded by the number of variables that can influence obesity including age, sex, diet, genetics, and environmental factors, to name but a few. Furthermore, it is possible that changes at the species level that cannot be detected by next-generation sequencing may be functionally relevant.
The intestinal microbiota has also been linked to heart disease in the presence of a high-fat diet (73). Increased metabolites of the dietary lipid phosphatidylcholine have been observed in the blood of patients suffering from myocardial infarction or stroke compared to that of normal individuals. In a mouse model, phosphatidylcholine metabolites such as choline are converted into trimethylamine by the intestinal microbiota. Trimethylamine is further metabolized in the liver to trimethylamine-N-oxide, which promotes arterial plaques and cardiovascular disease. These results raise the possibility that targeting particular components of the microbiota involved in choline metabolism may provide a novel therapeutic approach in the treatment of cardiovascular disease (67).
CANCER
Primary and sporadic mutations of tumor suppressor genes that accumulate in a characteristic progression underlie the development of cancer. Colorectal cancer provides a good model of this because the polyp to cancer progression can be followed visually and biologically. There is increasing evidence that inflammation may play a critical permissive role at all stages of cancer development and progression. Consequently, diseases that are characterized by chronic inflammation are often associated with a marked increase in cancer as is the case for colorectal carcinoma in IBD. Given the ability of the commensal microbiota to modulate inflammation, it is reasonable to consider whether the microbiota contributes to the pathogenesis of human carcinogenesis (74, 75). Among the cancers that have been linked to the microbiota are those involving the gastrointestinal tract and especially the colon where most of the microbes congregate, rather than the small intestine.
Sporadic colorectal cancer is typically initiated by somatic mutations in the adenomatous polyposis coli (APC) gene and when transmitted as a loss-of-function germ-line mutation is conferred in a Mendelian fashion and associated with early-onset familial colorectal cancer. Mice that are heterozygous for mutated Apc (ApcMin/+) are the most tractable model for studying gene-environment interactions in the development of colorectal cancer because they develop polyps upon loss of heterozygosity of the wild-type Apc allele and cancers upon the accumulation of additional mutations (76, 77). In contrast to ApcMin/+ mice with an intact microbiota, germ-free ApcMin/+ mice exhibit decreased numbers of adenomatous polyps (78). On the other hand, colonization of mice with enterotoxin-producing B. fragilis, a symbiote with pathogenic capabilities, which is often carried by asymptomatic individuals (79), can promote cancer development in ApcMin/+ mice. This occurs through β-catenin–driven epithelial cell proliferation by virtue of the toxin’s ability to degrade E-cadherin (80, 81). It is therefore possible that pathobionts and pathogens are important cofactors in the development of colorectal neoplasia. Whether this occurs through indirect (for example, effects on innate or adaptive immune functions even in the absence of overt inflammation or altered xenobiotic metabolism in conjunction with other environmental factors) and/or direct (for example, phenotypic exacerbation of an existing mutation or induction of a second genetic mutation) mechanisms is unknown. With regard to an indirect mechanism, ApcMin/+/Myd88−/− mice exhibit smaller tumors (82). With regard to a direct mechanism, Streptococcus gallolyticus ssp. gallolyticus, a biotype of Streptococcus bovis, which has been associated with colorectal cancer, can induce cyclooxygenase 2 expression (83). Cyclooxygenase 2 overexpression, which is often observed in colorectal cancer, is associated with a worse prognosis (84).
In humans, analysis of the microbiota in colon cancer tissue versus control tissue has also led to two independent reports of increased representation of Fusobacterium spp. and, in particular, Fusobacterium nucleatum, which has previously been associated with periodontal disease (85, 86). Whether these bacteria actually play a role in tumor development or progression or simply adapt to the intestinal cancer niche is not known. It has recently been suggested that particular members of the microbiota, such as B. fragilis discussed above, in addition to having virulence factors, may remodel the microbiome, favoring inflammatory responses that promote epithelial cell transformation, leading to cancer (87). This in turn may foster the emergence of additional microbes that might be considered as “carcinogenic allies” that further promote cancer progression through sustaining the inflammatory response and other mechanisms that directly affect the cancer cell. Such studies demonstrate how specific microbes may contribute to specific stages of colorectal cancer development perhaps in certain genetically susceptible contexts that are either encoded germ line or induced as a consequence of somatic or epigenetic alterations. This raises the possibility that eliminating such “pathogenic” organisms through specific antimicrobial measures, approaches that enable colonization resistance, or other means may be important avenues for the prevention of colorectal cancer and its development.
Such considerations also highlight the continuum that exists between chronic inflammation and the development of cancer. Inflammation-associated cancer has been observed in numerous animal models of inflammation and in people infected with inflammation-inducing microbial pathogens (for example, H. pylori) or in people with chronic inflammatory disorders like IBD (74, 75). H. pylori infection has been shown to promote both gastric carcinoma and mucosa-associated lymphoid tissue lymphoma (5). Because H. pylori is disappearing from human populations because of antibiotic treatment and reduced transmission, the incidence of gastric cancer has declined. In the case of IBD, inflammation-inducing pathobionts and loss of colonization resistance, as discussed above, may play a role in cancer development. For example, germ-free but not colonized mice deficient in interleukin-10, a key negative regulator of chronic inflammation and a genetic risk factor for primary and sporadic forms of human IBD, are resistant to chemically induced colorectal cancer (88). As previously hypothesized, infectious agents (and perhaps pathobionts) that are associated with neoplasia may drive tumorigenesis by their ability to promote proliferation, induce genetic errors, and suppress the ability of the immune system to eliminate the agent and/or transformed cells as modeled in viral systems (89).
It is also likely, however, that some components of the microbiota may prevent cancer (75). Thus, although the incidence of gastric cancer has decreased with the reduction in H. pylori infection, there has been an associated increase in gastroesophageal reflux disease and associated cancer. The mechanisms for this reciprocal relationship between gastric and esophageal cancer are not known but may relate to a change in the gastric microbiota in the absence of H. pylori with downstream effects on acid secretion, production of metabolic hormones, and the immune response (90). These studies suggest that changes in the microbiota in one location in the gastrointestinal tract (that is, the stomach) can affect cancer development at a different gastrointestinal site. Although defining the factors responsible for protection from carcinogenesis is of obvious benefit, these studies highlight the beneficial and detrimental roles played by the microbiota in the development of cancer.
IN PURSUIT OF NORMALITY AND REESTABLISHMENT OF HOMEOSTASIS
This discussion raises the question of whether the microbiota can be manipulated in a preventative and/or therapeutic manner in pursuit of defending against allergy and inflammation in a genetically susceptible host or stimulating tissue repair and restoring a durable state of homeostasis. To accomplish this will require an understanding of many different factors. Most important is the need to achieve a consensus definition of what comprises a normal microbiota and microbiome, and at each stage of life—a task that is challenged by the uniqueness of each individual in this regard. Accomplishing this also requires a deeper understanding of the biologic modules of the host to which the microbial components and metabolites are linked [see Review by Hooper et al. (91)]. Specifically, we are beginning to recognize that microorganisms and their expressed attributes are connected to specific biologic consequences, such as induction of distinct epithelial functions, secretory IgA immunity, and immune deviation of T cells to T helper 17 or T regulatory cells, for example (15, 91). With this knowledge, it can be envisioned that precise repair of induced or genetically based immune defects might be achievable by therapy with specific microbiota (or perhaps genetically engineered microbiota) or their functional components and/or prebiotics, which stimulate and sustain their expansion and persistence. An early example includes the administration of a Bifidobacterium lactis–containing milk product to mice genetically susceptible to colitis. The regimen promotes a microbial ecology that is associated with increased SCFA production and creation of an intestinal environment that prevents colonization with inflammation-inducing Enterobacteriaceae (92). Whether such approaches can be refined and translated to clinical practice such as humans with Crohn’s disease that are characterized by emergence of Enterobacteriaceae, including adherent-invasive E. coli, is unclear. Related types of fermented milk products have effected similar ecologic changes in human volunteers, however, raising the possibility of personalized approaches to repair of the microbiome (93).
At the other extreme would be those states in which the microbiome is completely “broken,” a meta-state that is also in need of a definition just as importantly as the requirement to explain normality. Just as in the case of organ failure wherein transplant is the final option, a movement is under way to similarly develop fecal transplantation as a therapeutic modality. Although recently demonstrated to be successful in an uncontrolled trial of 70 patients with chronic, relapsing C. difficile colitis, its long-term effects and consequences need to be understood (22).
Such studies illustrate the promise of manipulating the microbiome for the long-term benefit of humans in the prevention and treatment of chronic disease. Accomplishing this will require, among other things as discussed in the accompanying paper by Hooper and colleagues (91), improved mouse models, for example, that contain a humanized microbiota, which is characteristic of and phenocopies specific clinical conditions. Such models would allow for tractable therapeutic manipulations that would assist the testing and ultimately the translation of different potential therapeutic interventions (94).
Acknowledgments
We appreciate the helpful comments of A. Kaser (Cambridge), A. Onderdonk (Boston), and H. Uhlig (Oxford) and L. Paradiso for administrative assistance. Funding: R.B. was supported by NIH grants DK44319, DK51362, DK53056, and DK88199 and the Harvard Digestive Diseases Center (DK034854). F.P. is supported by a Wellcome Trust Senior Investigator Award.
REFERENCES AND NOTES
- 1.Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human–microbe mutualism and disease. Nature. 2007;449:811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dominguez-Bello MG, Blaser MJ, Ley RE, Knight R. Development of the human gastrointestinal microbiota and insights from high-throughput sequencing. Gastroenterology. 2011;140:1713–1719. doi: 10.1053/j.gastro.2011.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gonzalez A, Clemente JC, Shade A, Metcalf JL, Song S, Prithiviraj B, Palmer BE, Knight R. Our microbial selves: What ecology can teach us. EMBO Rep. 2011;12:775–784. doi: 10.1038/embor.2011.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474:327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blaser MJ, Falkow S. What are the consequences of the disappearing human microbiota? Nat. Rev. Microbiol. 2009;7:887–894. doi: 10.1038/nrmicro2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc. Natl. Acad. Sci. U.S.A. 2011;108(Suppl. 1):4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim SC, Tonkonogy SL, Albright CA, Tsang J, Balish EJ, Braun J, Huycke MM, Sartor RB. Variable phenotypes of enterocolitis in interleukin 10-deficient mice mono-associated with two different commensal bacteria. Gastroenterology. 2005;128:891–906. doi: 10.1053/j.gastro.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 9.Lees CW, Barrett JC, Parkes M, Satsangi J. New IBD genetics: Common pathways with other diseases. Gut. 2011;60:1739–1753. doi: 10.1136/gut.2009.199679. [DOI] [PubMed] [Google Scholar]
- 10.Holmes E, Li JV, Athanasiou T, Ashrafian H, Nicholson JK. Understanding the role of gut microbiome–host metabolic signal disruption in health and disease. Trends Microbiol. 2011;19:349–359. doi: 10.1016/j.tim.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 11.Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: Evolution of the vertebrate gut microbiota. Nat. Rev. Microbiol. 2008;6:776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neish AS. Microbes in gastrointestinal health and disease. Gastroenterology. 2009;136:65–80. doi: 10.1053/j.gastro.2008.10.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mushin R, Dubos R. Colonization of the mouse intestine with Escherichia coli. J. Exp. Med. 1965;122:745–757. doi: 10.1084/jem.122.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stecher B, Hardt WD. The role of microbiota in infectious disease. Trends Microbiol. 2008;16:107–114. doi: 10.1016/j.tim.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 15.Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat. Rev. Immunol. 2010;10:159–169. doi: 10.1038/nri2710. [DOI] [PubMed] [Google Scholar]
- 16.Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho JH, Murray TS, Iwasaki A. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc. Natl. Acad. Sci. U.S.A. 2011;108:5354–5359. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamada N, Kim YG, Sham HP, Vallance BA, Puente JL, Martens EC, Núñez G. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science. 2012 doi: 10.1126/science.1222195. 10.1126/science.1222195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guina T, Yi EC, Wang H, Hackett M, Miller SI. A PhoP-regulated outer membrane protease of Salmonella enterica serovar typhimurium promotes resistance to alpha-helical antimicrobial peptides. J. Bacteriol. 2000;182:4077–4086. doi: 10.1128/jb.182.14.4077-4086.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ubeda C, Taur Y, Jenq RR, Equinda MJ, Son T, Samstein M, Viale A, Socci ND, van den Brink MR, Kamboj M, Pamer EG. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J. Clin. Invest. 2010;120:4332–4341. doi: 10.1172/JCI43918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bartlett JG. Narrative review: The new epidemic of Clostridium difficile–associated enteric disease. Ann. Intern. Med. 2006;145:758–764. doi: 10.7326/0003-4819-145-10-200611210-00008. [DOI] [PubMed] [Google Scholar]
- 22.Mattila E, Uusitalo-Seppälä R, Wuorela M, Lehtola L, Nurmi H, Ristikankare M, Moilanen V, Salminen K, Seppälä M, Mattila PS, Anttila VJ, Arkkila P. Fecal transplantation, through colonoscopy, is effective therapy for recurrent Clostridium difficile infection. Gastroenterology. 2012;142:490–496. doi: 10.1053/j.gastro.2011.11.037. [DOI] [PubMed] [Google Scholar]
- 23.Manary MJ, Heikens GT, Golden M. Kwashiorkor: More hypothesis testing is needed to understand the aetiology of oedema. Malawi Med. J. 2009;21:106–107. doi: 10.4314/mmj.v21i3.45630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, Finlay BB. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2:119–129. doi: 10.1016/j.chom.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 25.van der Waaij D. The ecology of the human intestine and its consequences for overgrowth by pathogens such as Clostridium difficile. Annu. Rev. Microbiol. 1989;43:69–87. doi: 10.1146/annurev.mi.43.100189.000441. [DOI] [PubMed] [Google Scholar]
- 26.Enoch DA, Butler MJ, Pai S, Aliyu SH, Karas JA. Clostridium difficile in children: Colonisation and disease. J. Infect. 2011;63:105–113. doi: 10.1016/j.jinf.2011.05.016. [DOI] [PubMed] [Google Scholar]
- 27.Medzhitov R, Schneider DS, Soares MP. Disease tolerance as a defense strategy. Science. 2012;335:936–941. doi: 10.1126/science.1214935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaser A, Zeissig S, Blumberg RS. Inflammatory bowel disease. Annu. Rev. Immunol. 2010;28:573–621. doi: 10.1146/annurev-immunol-030409-101225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cho I, Blaser MJ. The human microbiome: At the interface of health and disease. Nat. Rev. Genet. 2012;13:260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raoult D. The globalization of intestinal microbiota. Eur. J. Clin. Microbiol. Infect. Dis. 2010;29:1049–1050. doi: 10.1007/s10096-010-0977-0. [DOI] [PubMed] [Google Scholar]
- 31.Turnbaugh PJ, Quince C, Faith JJ, McHardy AC, Yatsunenko T, Niazi F, Affourtit J, Egholm M, Henrissat B, Knight R, Gordon JI. Organismal, genetic, and transcriptional variation in the deeply sequenced gut microbiomes of identical twins. Proc. Natl. Acad. Sci. U.S.A. 2010;107:7503–7508. doi: 10.1073/pnas.1002355107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Strober W, Fuss IJ, Blumberg RS. The immunology of mucosal models of inflammation. Annu. Rev. Immunol. 2002;20:495–549. doi: 10.1146/annurev.immunol.20.100301.064816. [DOI] [PubMed] [Google Scholar]
- 33.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by Toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 34.Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, Schilter HC, Rolph MS, Mackay F, Artis D, Xavier RJ, Teixeira MM, Mackay CR. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Olszak T, An D, Zeissig S, Vera MP, Richter J, Fanke A, Glickman JN, Siebert R, Baron RM, Kasper DL, Blumberg RS. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–493. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134:577–594. doi: 10.1053/j.gastro.2007.11.059. [DOI] [PubMed] [Google Scholar]
- 37.Shaw SY, Blanchard JF, Bernstein CN. Association between the use of antibiotics in the first year of life and pediatric inflammatory bowel disease. Am. J. Gastroenterol. 2010;105:2687–2692. doi: 10.1038/ajg.2010.398. [DOI] [PubMed] [Google Scholar]
- 38.Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE. Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl. Acad. Sci. U.S.A. 2011;108(Suppl. 1):4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Renz H, Brandtzaeg P, Hornef M. The impact of perinatal immune development on mucosal homeostasis and chronic inflammation. Nat. Rev. Immunol. 2011;12:9–23. doi: 10.1038/nri3112. [DOI] [PubMed] [Google Scholar]
- 40.Chassaing B, Darfeuille-Michaud A. The commensal microbiota and enteropathogens in the pathogenesis of inflammatory bowel diseases. Gastroenterology. 2011;140:1720–1728. doi: 10.1053/j.gastro.2011.01.054. [DOI] [PubMed] [Google Scholar]
- 41.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, Gratadoux JJ, Blugeon S, Bridonneau C, Furet JP, Corthier G, Grangette C, Vasquez N, Pochart P, Trugnan G, Thomas G, Blottière HM, Doré J, Marteau P, Seksik P, Langella P. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. U.S.A. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307–317. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maloy KJ, Powrie F. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature. 2011;474:298–306. doi: 10.1038/nature10208. [DOI] [PubMed] [Google Scholar]
- 44.Kobayashi KS, Chamaillard M, Ogura Y, Henegariu O, Inohara N, Nuñez G, Flavell RA. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 2005;307:731–734. doi: 10.1126/science.1104911. [DOI] [PubMed] [Google Scholar]
- 45.Rehman A, Sina C, Gavrilova O, Häsler R, Ott S, Baines JF, Schreiber S, Rosenstiel P. Nod2 is essential for temporal development of intestinal microbial communities. Gut. 2011;60:1354–1362. doi: 10.1136/gut.2010.216259. [DOI] [PubMed] [Google Scholar]
- 46.Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, Ley R, Wakeland EK, Hooper LV. The antibacterial lectin RegIIIγ promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, Kishi C, Kc W, Carrero JA, Hunt S, Stone CD, Brunt EM, Xavier RJ, Sleckman BP, Li E, Mizushima N, Stappenbeck TS, Virgin HW., IV A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cadwell K, Patel KK, Maloney NS, Liu TC, Ng AC, Storer CE, Head RD, Xavier R, Stappenbeck TS, Virgin HW. Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141:1135–1145. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peterson DA, Frank DN, Pace NR, Gordon JI. Metagenomic approaches for defining the pathogenesis of inflammatory bowel diseases. Cell Host Microbe. 2008;3:417–427. doi: 10.1016/j.chom.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. U.S.A. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Willing BP, Dicksved J, Halfvarson J, Andersson AF, Lucio M, Zheng Z, Järnerot G, Tysk C, Jansson JK, Engstrand L. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology. 2010;139:1844.e1–1854.e1. doi: 10.1053/j.gastro.2010.08.049. [DOI] [PubMed] [Google Scholar]
- 52.Lepage P, Häsler R, Spehlmann ME, Rehman A, Zvirbliene A, Begun A, Ott S, Kupcinskas L, Doré J, Raedler A, Schreiber S. Twin study indicates loss of interaction between microbiota and mucosa of patients with ulcerative colitis. Gastroenterology. 2011;141:227–236. doi: 10.1053/j.gastro.2011.04.011. [DOI] [PubMed] [Google Scholar]
- 53.Willing B, Halfvarson J, Dicksved J, Rosenquist M, Järnerot G, Engstrand L, Tysk C, Jansson JK. Twin studies reveal specific imbalances in the mucosa-associated microbiota of patients with ileal Crohn’s disease. Inflamm. Bowel Dis. 2009;15:653–660. doi: 10.1002/ibd.20783. [DOI] [PubMed] [Google Scholar]
- 54.Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C, Davies J, Ervine A, Poulter L, Pachter L, Moffatt MF, Cookson WO. Disordered microbial communities in asthmatic airways. PLoS One. 2010;5:e8578. doi: 10.1371/journal.pone.0008578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maldonado-Contreras A, Goldfarb KC, Godoy-Vitorino F, Karaoz U, Contreras M, Blaser MJ, Brodie EL, Dominguez-Bello MG. Structure of the human gastric bacterial community in relation to Helicobacter pylori status. ISME J. 2011;5:574–579. doi: 10.1038/ismej.2010.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, Peaper DR, Bertin J, Eisenbarth SC, Gordon JI, Flavell RA. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, Delaney ML, Punit S, Karlsson M, Bry L, Glickman JN, Gordon JI, Onderdonk AB, Glimcher LH. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bloom SM, Bijanki VN, Nava GM, Sun L, Malvin NP, Donermeyer DL, Dunne WM, Jr, Allen PM, Stappenbeck TS. Commensal Bacteroides species induce colitis in host-genotype-specific fashion in a mouse model of inflammatory bowel disease. Cell Host Microbe. 2011;9:390–403. doi: 10.1016/j.chom.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Strachan DP. Hay fever, hygiene, and household size. BMJ. 1989;299:1259–1260. doi: 10.1136/bmj.299.6710.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Okada H, Kuhn C, Feillet H, Bach JF. The ‘hygiene hypothesis’ for autoimmune and allergic diseases: An update. Clin. Exp. Immunol. 2010;160:1–9. doi: 10.1111/j.1365-2249.2010.04139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ege MJ, Mayer M, Normand AC, Genuneit J, Cookson WO, Braun-Fahrländer C, Heederik D, Piarroux R, von Mutius E GABRIELA Transregio 22 Study Group. Exposure to environmental microorganisms and childhood asthma. N. Engl. J. Med. 2011;364:701–709. doi: 10.1056/NEJMoa1007302. [DOI] [PubMed] [Google Scholar]
- 62.Arnold IC, Dehzad N, Reuter S, Martin H, Becher B, Taube C, Müller A. Helicobacter pylori infection prevents allergic asthma in mouse models through the induction of regulatory T cells. J. Clin. Invest. 2011;121:3088–3093. doi: 10.1172/JCI45041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Troy EB, Kasper DL. Beneficial effects of Bacteroides fragilis polysaccharides on the immune system. Front. Biosci. 2010;15:25–34. doi: 10.2741/3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Russell SL, Gold MJ, Hartmann M, Willing BP, Thorson L, Wlodarska M, Gill N, Blanchet MR, Mohn WW, McNagny KM, Finlay BB. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep. 2012;13:440–447. doi: 10.1038/embor.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hill DA, Siracusa MC, Abt MC, Kim BS, Kobuley D, Kubo M, Kambayashi T, Larosa DF, Renner ED, Orange JS, Bushman FD, Artis D. Commensal bacteria-derived signals regulate basophil hematopoiesis and allergic inflammation. Nat. Med. 2012;18:538–546. doi: 10.1038/nm.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Holmes E, Kinross J, Gibson GR, Burcelin R, Jia W, Pettersson S, Nicholson JK. Therapeutic modulation of microbiota-host metabolic interactions. Sci. Transl. Med. 2012;4:137rv6. doi: 10.1126/scitranslmed.3004244. [DOI] [PubMed] [Google Scholar]
- 67.Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 68.Samuel BS, Shaito A, Motoike T, Rey FE, Backhed F, Manchester JK, Hammer RE, Williams SC, Crowley J, Yanagisawa M, Gordon JI. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proc. Natl. Acad. Sci. U.S.A. 2008;105:16767–16772. doi: 10.1073/pnas.0808567105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, Sitaraman SV, Knight R, Ley RE, Gewirtz AT. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 71.Shulzhenko N, Morgun A, Hsiao W, Battle M, Yao M, Gavrilova O, Orandle M, Mayer L, Macpherson AJ, McCoy KD, Fraser-Liggett C, Matzinger P. Crosstalk between B lymphocytes, microbiota and the intestinal epithelium governs immunity versus metabolism in the gut. Nat. Med. 2011;17:1585–1593. doi: 10.1038/nm.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Korecka A, Arulampalam V. The gut microbiome: Scourge, sentinel or spectator? J. Oral Microbiol. 2012;4 doi: 10.3402/jom.v4i0.9367. 10.3402/jom.v4i0.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, Feldstein AE, Britt EB, Fu X, Chung YM, Wu Y, Schauer P, Smith JD, Allayee H, Tang WH, DiDonato JA, Lusis AJ, Hazen SL. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Boleij A, Tjalsma H. Gut bacteria in health and disease: A survey on the interface between intestinal microbiology and colorectal cancer. Biol. Rev. Camb. Philos. Soc. 2012 doi: 10.1111/j.1469-185X.2012.00218.x. 10.1111/j.1469-185X.2012.00218.x. [DOI] [PubMed] [Google Scholar]
- 76.Lee SH, Hu LL, Gonzalez-Navajas J, Seo GS, Shen C, Brick J, Herdman S, Varki N, Corr M, Lee J, Raz E. ERK activation drives intestinal tumorigenesis in Apcmin/+ mice. Nat. Med. 2010;16:665–670. doi: 10.1038/nm.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yamada Y, Mori H. Multistep carcinogenesis of the colon in ApcMin/+ mouse. Cancer Sci. 2007;98:6–10. doi: 10.1111/j.1349-7006.2006.00348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dove WF, Clipson L, Gould KA, Luongo C, Marshall DJ, Moser AR, Newton MA, Jacoby RF. Intestinal neoplasia in the ApcMin mouse: Independence from the microbial and natural killer (beige locus) status. Cancer Res. 1997;57:812–814. [PubMed] [Google Scholar]
- 79.Sears CL. Enterotoxigenic Bacteroides fragilis: A rogue among symbiotes. Clin. Microbiol. Rev. 2009;22:349–369. doi: 10.1128/CMR.00053-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wu S, Rhee KJ, Zhang M, Franco A, Sears CL. Bacteroides fragilis toxin stimulates intestinal epithelial cell shedding and γ-secretase-dependent E-cadherin cleavage. J. Cell Sci. 2007;120(Pt. 11):1944–1952. doi: 10.1242/jcs.03455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Toprak NU, Yagci A, Gulluoglu BM, Akin ML, Demirkalem P, Celenk T, Soyletir G. A possible role of Bacteroides fragilis enterotoxin in the aetiology of colorectal cancer. Clin. Microbiol. Infect. 2006;12:782–786. doi: 10.1111/j.1469-0691.2006.01494.x. [DOI] [PubMed] [Google Scholar]
- 82.Rakoff-Nahoum S, Medzhitov R. Regulation of spontaneous intestinal tumorigenesis through the adaptor protein MyD88. Science. 2007;317:124–127. doi: 10.1126/science.1140488. [DOI] [PubMed] [Google Scholar]
- 83.Abdulamir AS, Hafidh RR, Abu Bakar F. The association of Streptococcus bovis/gallolyticus with colorectal tumors: The nature and the underlying mechanisms of its etiological role. J. Exp. Clin. Cancer Res. 2011;30:11. doi: 10.1186/1756-9966-30-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ogino S, Kirkner GJ, Nosho K, Irahara N, Kure S, Shima K, Hazra A, Chan AT, Dehari R, Giovannucci EL, Fuchs CS. Cyclooxygenase-2 expression is an independent predictor of poor prognosis in colon cancer. Clin. Cancer Res. 2008;14:8221–8227. doi: 10.1158/1078-0432.CCR-08-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, Ojesina AI, Jung J, Bass AJ, Tabernero J, Baselga J, Liu C, Shivdasani RA, Ogino S, Birren BW, Huttenhower C, Garrett WS, Meyerson M. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, Barnes R, Watson P, Allen-Vercoe E, Moore RA, Holt RA. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sears CL, Pardoll DM. Perspective: Alpha-bugs, their microbial partners, and the link to colon cancer. J. Infect. Dis. 2011;203:306–311. doi: 10.1093/jinfdis/jiq061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Uronis JM, Jobin C. Microbes and colorectal cancer: Is there a relationship? Curr. Oncol. 2009;16:22–24. doi: 10.3747/co.v16i4.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Klein G. Lymphoma development in mice and humans: Diversity of initiation is followed by convergent cytogenetic evolution. Proc. Natl. Acad. Sci. U.S.A. 1979;76:2442–2446. doi: 10.1073/pnas.76.5.2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Plottel CS, Blaser MJ. Microbiome and malignancy. Cell Host Microbe. 2011;10:324–335. doi: 10.1016/j.chom.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Veiga P, Gallini CA, Beal C, Michaud M, Delaney ML, DuBois A, Khlebnikov A, van Hylckama Vlieg JE, Punit S, Glickman JN, Onderdonk A, Glimcher LH, Garrett WS. Bifidobacterium animalis subsp. lactis fermented milk product reduces inflammation by altering a niche for colitogenic microbes. Proc. Natl. Acad. Sci. U.S.A. 2010;107:18132–18137. doi: 10.1073/pnas.1011737107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.McNulty NP, Yatsunenko T, Hsiao A, Faith JJ, Muegge BD, Goodman AL, Henrissat B, Oozeer R, Cools-Portier S, Gobert G, Chervaux C, Knights D, Lozupone CA, Knight R, Duncan AE, Bain JR, Muehlbauer MJ, Newgard CB, Heath AC, Gordon JI. The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Sci. Transl. Med. 2011;3:106ra106. doi: 10.1126/scitranslmed.3002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Reading NC, Kasper DL. The starting lineup: Key microbial players in intestinal immunity and homeostasis. Front. Microbiol. 2011;2:148. doi: 10.3389/fmicb.2011.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]