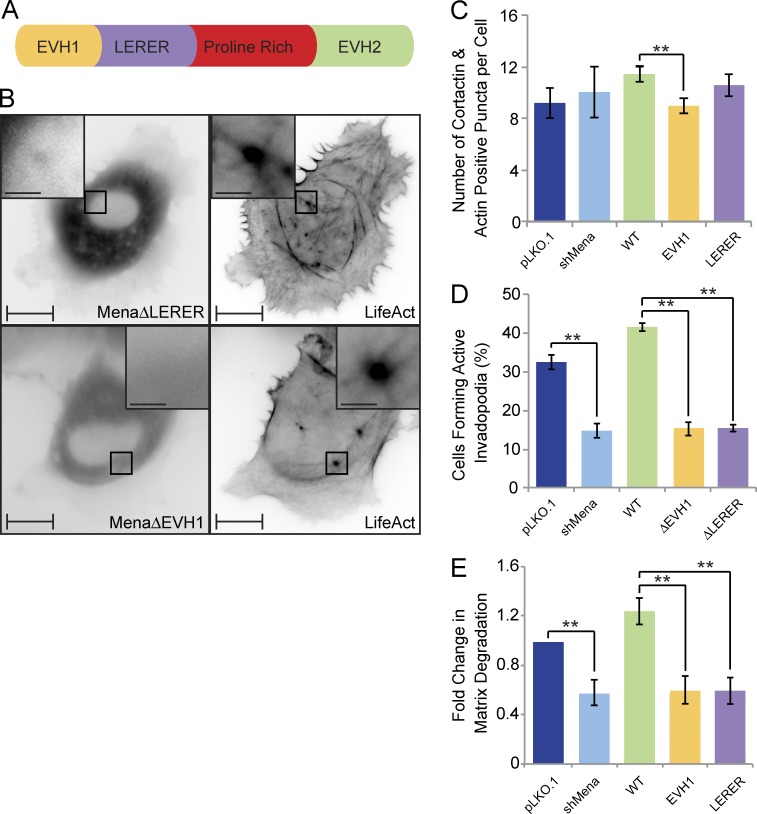
Figure 7.
Mena is recruited to invadopodia via its EVH1 domain and requires the LERER sequences for its ring-like localization and proteolytic activity. (A) Schematic diagram of Mena depicts its EVH1 domain, LERER repeats, proline-rich region, and the EVH2 domain. (B) MDA-MB-231-shMena cells were transiently transfected with tagRFP-LifeAct and GFP-MenaΔEVH1 or GFP-MenaΔLERER and subjected to time-lapse video microscopy. Single frame illustrates GFP-MenaΔEVH1 and GFP-MenaΔLERER localization with respect to F-actin cores of invadopodia protrusions. (C) MDA-MB-231-pLKO.1, MDA-MB-231-shMena, and MDA-MB-231-shMena cells transiently rescued with WT-Mena, MenaΔEVH1, or MenaΔLERER were seeded on unconjugated gelatin matrix and immunostained for invadopodia markers (F-actin and cortactin), and confocal images were acquired at the ventral cell surface. The number of F-actin– and cortactin-positive protrusions per cell was quantified. (D) MDA-MB-231-pLKO.1, MDA-MB-231-shMena, and MDA-MB-231-shMena cells transiently rescued with WT-Mena, MenaΔEVH1, or MenaΔLERER were seeded on fluorescent gelatin matrix and immunostained with phalloidin (F-actin), confocal images were acquired at the ventral cell surface, and the percentage of cells that formed proteolytically active invadopodia (positive for F-actin punctate overlaying matrix degradation area) was quantified. (E) As in D, invadopodia assay was performed, and the amount of ECM degradation (mean matrix degradation area per fields of view) by MDA-MB-231-pLKO.1, MDA-MB-231-shMena, and MDA-MB-231-shMena cells transiently rescued with WT-Mena, MenaΔEVH1, or MenaΔLERER was quantified as fold change compared with control (pLKO.1) cells. All quantified data indicate the mean values ± SE from at least three independent experiments. **, P < 0.01. Bars: 10 µm; (inset) 2 µm.