Figure 6.
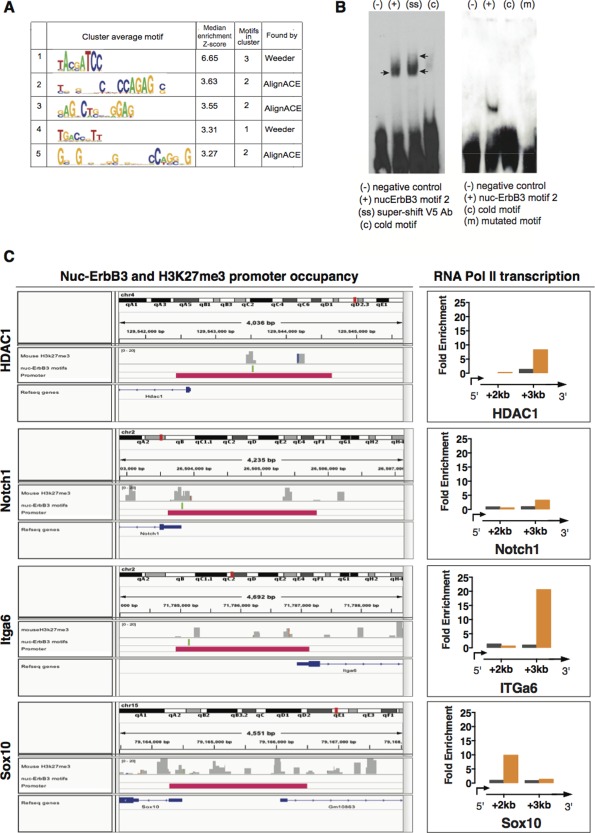
(A) Nuc‐ErbB3 binding motifs were located from ChIP‐chip peak data using the program WebMotifs, which combines three de novo motif discovery programs: AlignACE, Weeder and MDscan. 5 groups of motifs were identified. (B) EMSA with nuclear extracts from rat Schwann cells transfected with nuc‐ErbB3‐V5 show specific binding to motif 2. Addition of anti‐V5 antibody results in a supershift (arrows). Cold (nonbiotinylated) oligo competes and prevents binding of motif 2 to nuc‐ErbB3 (left panel). EMSA with recombinant nuc‐ErbB3 shows specific binding to motif 2. Cold (nonbiotinylated) oligo competes and prevents binding of motif 2 to nuc‐ErbB3, while mutation of the core sequence of motif 2 abrogates binding to recombinant nuc‐ErbB3 (right panel). (C) Computational analysis of nuc‐ErbB3 and H3K27me3 promoter occupancy from in vivo sciatic nerve ChIP data. Co‐occurrence of nuc‐ErbB3 binding motifs and chromatin regions associated with H3K27me3 were discovered in the promoters of HDAC1, Notch1 and Itga6 while Sox10 promoter contains only H3K27me3 binding sites. Sonicated chromatin isolated from P30 sciatic nerves of WT or nuc‐ErbB3 KI mice was immunoprecipitated with antibodies against RNA Polymerase II (Millipore). Fold enrichment over input is presented for HDAC, Notch1, Itga6, and Sox10. Localization of RNA Pol II greater than 1 kb downstream of the TSS identifies active transcription and increased levels at the nuc‐ErbB3 KI samples indicate elevated transcriptional rate following loss of nuc‐ErbB3 from the nucleus. Genes with nuc‐ErbB3 and H3K27me3 co‐occupancy on their promoters (e.g., HDAC1, Itga6, and Notch1) are transcriptionally repressed since they exhibit increased rate of transcription (derepression) in the absence of nuc‐ErbB3 in KI mice. The same effect on transcription was observed for Sox10 verifying that the association of nuc‐ErbB3 with Sox10 promoter, which occurs in the context of the repressive histone mark H3K27me3 without direct DNA binding, results in transcriptional repression. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
