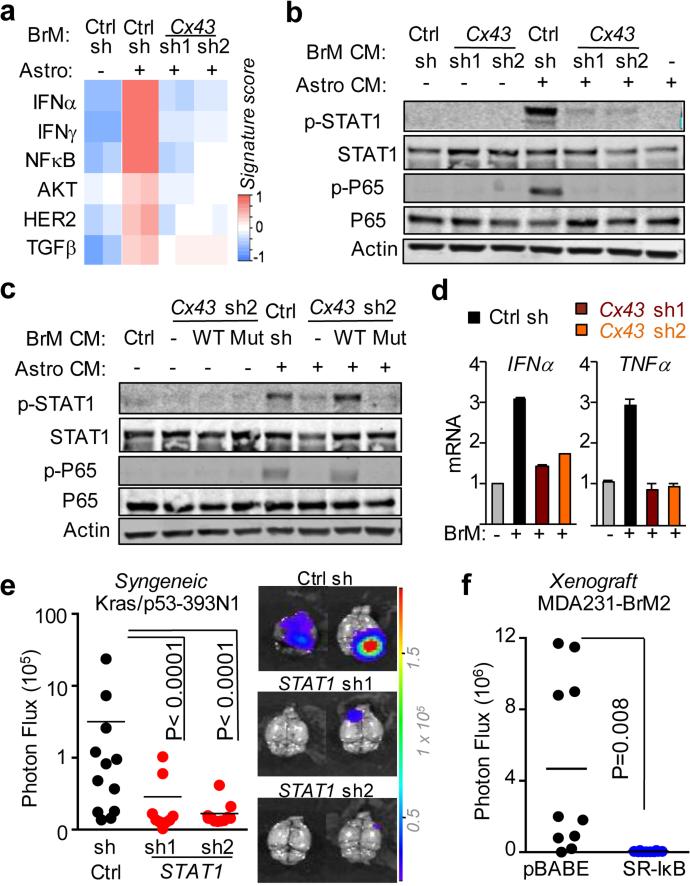
Figure 3. Gap junctions activate STAT1 and NF-κB pathways in cancer cells.
a, Signaling pathway analysis of TRAP-Seq data from MDA231-BrM2 cells after co-culture with astrocytes. Control (Ctrl) or Cx43-depleted MDA231-BrM2 cells expressing an L10a-GFP ribosomal protein fusion were co-cultured with astrocytes for 24 h prior to polysome immunoprecipitation and mRNA sequencing. Heatmap depicts blue (down-regulated) and red (up-regulated) pathways. n=2 biological replicates. b,c, STAT1 and NF-κB p65 phosphorylation in MDA231-BrM2 cells after a 2 h incubation with conditioned media (CM) from astrocyte co-culture. CM were collected after 24 h co-culture of astrocytes with control or Cx43-depleted MDA231-BrM2 cells (b), or from Cx43-depleted MDA231-BrM2 cells that were transduced with wild type Cx43 (WT) or Cx43(T154A) mutant (Mut) (c), n≥3 independent experiments both (b) and (c). d, Relative mRNA levels of IFNA and TNFA in human astrocytes re-isolated after co-culture with MDA231-BrM2 cancer cells. All values are mean ± S.E.M. (Data are n=3 biological replicates in 2 independent experiments). e-f, Quantification of BLI signal from brain metastases formed by control or STAT1-knockdown Kras/p53-393N1 cells (e), and SR-IκBα MDA231-BrM2 cells (f) (Data are from n=2 independent experiments, with 12 mice total per group).