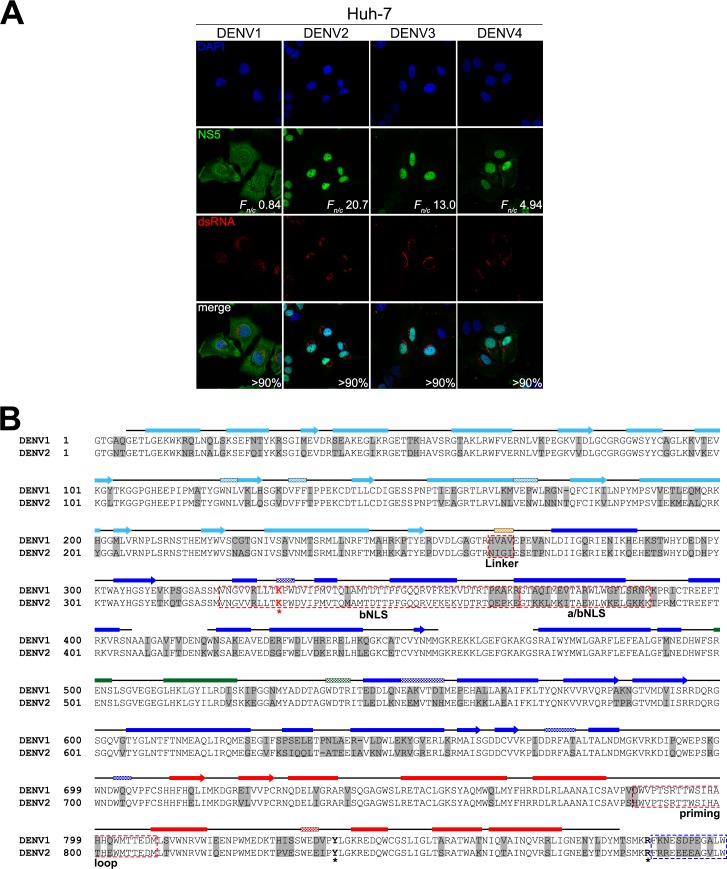
Fig 1. Subcellular localization of NS5 in DENV1-4 infected cells and sequence alignment of DENV1 and 2 NS5.
(A) Huh-7 cells were infected with DENV1-4 at MOI 10 and the infected cells (>90%) were analysed for presence of NS5 (green) and dsRNA (red) by IFA at 24h post-infection. Digitized images were captured by Zeiss LSM 710 upright confocal microscope by 63× oil immersion lens. Image analysis was performed on digitized images of NS5 staining with ImageJ software [52] to determine nuclear to cytoplasmic fluorescence ratio (F n/c) as done previously [17, 29, 30, 42]. (B) Sequence and structural analysis of DENV1 NS5 aligned against DENV2 NS5. The alignment was performed using Clustal Omega and the numbering of the alignment is based on DENV2 NS5. The PDB file of DENV3 NS5 protein (PDB: 4V0Q, [34] was used as the input file for secondary structure depiction. The secondary structures are indicated by solid boxes (α-helices), checked boxes (310 helix) and arrows (β-sheets) above the sequence alignment and they are coloured light blue for the Mtase domain, orange for linker (residues 264–273) and blue for the fingers, green for the palm and red for the thumb of the RdRp domain. The bNLS sequence (residues 320–368) [31], the a/bNLS sequence (residues 369–389) [29, 30] and the priming loop sequence (residues 786–809) [13] are boxed in red. Within bNLS, K330 (red asterisk and bold) is important for NS3–NS5 interaction [26]. NS5 residues that are not identical to DENV1 and DENV2 are highlighted in grey. Other highlighted residues Y838 and R888 are discussed in the text. The electron density of the last 12 amino acid residues (boxed in blue) are generally missing in most DENV NS5 crystal structures (PDB: 2J7U, 4C11 and 4V0Q). The GenBank accession numbers of DENV1 and DENV2 are EU081230 and EU081177, respectively.