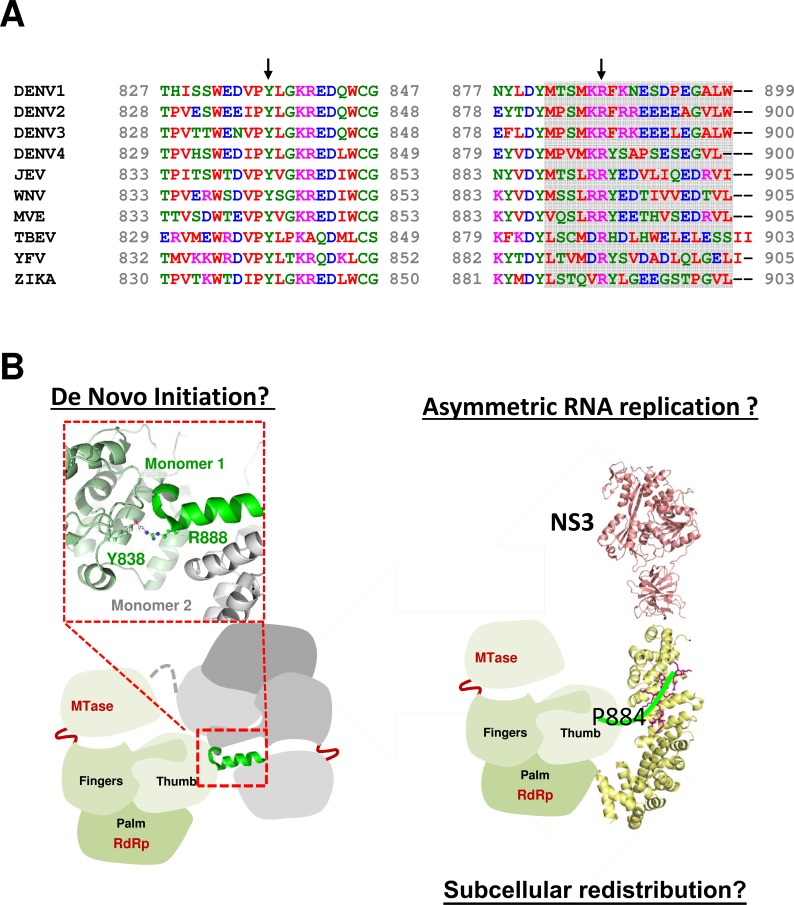
Fig 9. Sequence variation and conformational flexibility of NS5 Cter18 regulates the oligomeric state, nuclear localization, viral replication.
(A) Sequence variation of residues 828–848 and 883–900 of DENV1–4 and representative flaviviruses. Arrow highlights the completely conserved Y838 and R888. The alignment was performed using Clustal Omega. The virus sequences and their GenBank accession numbers are as follows: DENV1-4 (same GenBank accession number as above), Japanese Encephalitis virus (JEV; M55506), West Nile virus (WNV; M12294), Murray Valley Encephalitis virus (MVE; AF161266), Tick-Borne Encephalitis Virus (TBEV; U27495), Yellow Fever virus (YFV; X15062) and Zika virus (ZIKV;KU497555) [58]. (B) Model of the role of the C-terminal region of NS5 that is required for the formation of dimer and the initiation of de novo viral RNA replication and interaction with importin-α or NS3. On the left: de novo RNA synthesis by NS5 oligomers independent of NS3 interaction as implied by NS3 N570A mutation [59] and non-viability of NS5 R888A as identified in this study. On the right: NS3 interaction with NS5 thumb domain in the RC will support higher plus strand RNA synthesis or NS5 Cter18 interacts with host importins or other factors to be localized to nucleus or cytoplasm in a regulated manner.