Abstract
TFIIH is a 10-subunit RNA polymerase II basal transcription factor with a dual role in DNA repair. TFIIH contains three enzymatic functions and over 30 conserved subdomains and topological regions. We systematically tested the function of these regions in three TFIIH core module subunits, i.e., Ssl1, Tfb4, and Tfb2, in the DNA translocase subunit Ssl2, and in the kinase module subunit Tfb3. Our results are consistent with previously predicted roles for the Tfb2 Hub, Ssl2 Lock, and Tfb3 Latch regions, with mutations in these elements typically having severe defects in TFIIH subunit association. We also found unexpected roles for other domains whose function had not previously been defined. First, the Ssl1-Tfb4 Ring domains are important for TFIIH assembly. Second, the Tfb2 Hub and HEAT domains have an unexpected role in association with Tfb3. Third, the Tfb3 Ring domain is important for association with many other TFIIH subunits. Fourth, a partial deletion of the Ssl1 N-terminal extension (NTE) domain inhibits TFIIH function without affecting subunit association. Finally, we used site-specific cross-linking to localize the Tfb3-binding surface on the Rad3 Arch domain. Our cross-linking results suggest that Tfb3 and Rad3 have an unusual interface, with Tfb3 binding on two opposite faces of the Arch.
INTRODUCTION
TFIIH is a conserved 10-subunit complex that plays essential roles in both RNA polymerase II (Pol II) transcription and nucleotide excision repair (NER) (1–3). TFIIH and TFIIE are the last basal factors to join the transcription preinitiation complex (PIC) (4). In transcription, TFIIH functions in DNA unwinding, transcription start site (TSS) scanning, promoter escape, and Pol II C-terminal domain (CTD) phosphorylation. During NER, TFIIH promotes the DNA unwinding of an asymmetric region around the site of DNA lesions as an early step in excision and replacement of damaged DNA (5, 6). Mutations in three TFIIH subunits, XPD, XPB, and p8, cause human diseases associated with defects in DNA repair and transcription (1).
TFIIH has three enzymatic activities that are essential for these functions. First, the Rad3/XPD (yeast/human) subunit is an ATP-dependent DNA helicase that participates in DNA unwinding during NER. While this enzymatic activity is dispensable for transcription, Rad3/XPD is required for transcription as it plays an essential structural role in stabilizing the TFIIH complex (7–10). Second, Ssl2/XPB is an ATP-dependent double-stranded DNA translocase with roles in transcription and DNA repair. Ssl2 promotes formation of the Pol II open complex by reeling downstream promoter DNA into the Pol II active site, leading to DNA unwinding (11, 12). In budding yeast, Pol II transcription initiates downstream from the site of PIC formation (13), and Ssl2 function drives the scanning of downstream promoter DNA for an appropriate TSS (14). In the human transcription system, XPB activity has been implicated in promoter escape and the release of Pol II from contacts with promoter DNA and other PIC components (15, 16). Ssl2/XPB activity also cooperates with Rad3/XPD in forming the initial DNA bubble surrounding DNA lesions (5). Third, three subunits of TFIIH (yeast Kin28/Ccl1/Tfb3) form a kinase module that targets the Pol II CTD for phosphorylation at Ser5 and Ser7 (1). In vivo, Ser5 phosphorylation plays a role in promoter escape as well as mRNA capping and other early steps in elongation. In mammals, this three-subunit complex, termed cyclin-dependent kinase (CDK)-activating kinase (CAK), also functions in cell cycle control by phosphorylation of at least four CDKs involved in cell cycle regulation (17). During NER, the kinase module is thought to dissociate from TFIIH to allow binding of the repair factor XPA (18).
Because of its flexible structure and complex subunit composition, the architecture of TFIIH is the least understood of the Pol II basal factors. Low-resolution electron microscopy (EM) structures of yeast and human TFIIH have been generated from complete and partial TFIIH complexes and TFIIH-containing PICs (19–23). A combination of structural and biochemical studies showed that TFIIH interacts with promoter DNA downstream from the initial site of DNA unwinding via the Ssl2/XPB subunit (12, 22–25). In the PIC, TFIIH also interacts with TFIIE and the Med11 subunit of the Mediator coactivator complex (12, 26, 27).
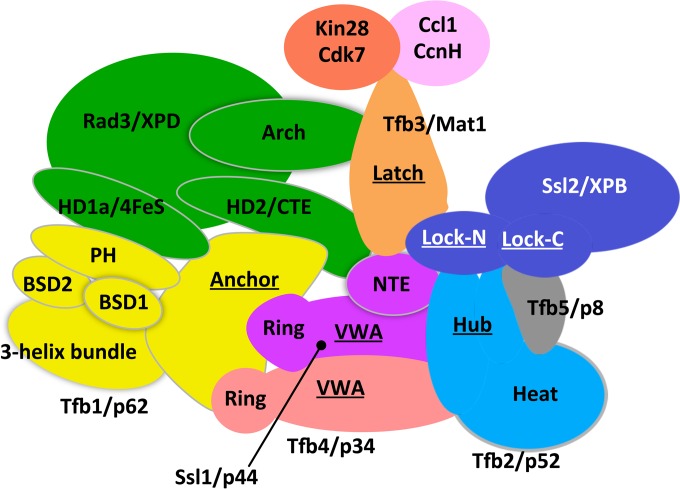
Biochemical analysis showed that TFIIH contains a set of core subunits consisting of Rad3/XPD, Tfb1/p62, Tfb2/p44, Tfb4/p34, and Tfb5/p8 (1, 21, 28). The translocase Ssl2/XPB reversibly dissociates from the core, with dissociation promoted by the factor Tfb6 (29). The three-subunit kinase module dissociates during NER as noted above. There are over 30 predicted and experimentally determined structured domains within TFIIH subunits, and many studies have examined intermolecular interactions between these subunits (summarized in reference 30). Recently, we probed the architecture of yeast and human TFIIH using chemical cross-linking mass spectrometry (MS) in combination with structural, genetic, and biochemical information (30). This study identified four new topological regions within TFIIH (the Tfb1 Anchor, the Tfb2 Hub, the Tfb3 Latch, and the Ssl2 Lock) that were predicted to organize many of the TFIIH subunit-subunit interactions and provided an initial framework for understanding TFIIH functions (Fig. 1).
FIG 1.
Model of TFIIH subunit organization and subdomain interactions based on cross-linking–mass spectrometry (adapted from reference 30 with permission of the publisher).
Here, we report the results of a systematic analysis to analyze the function of five TFIIH subunits. Using a yeast system that allows testing the genetic and biochemical phenotypes of TFIIH subunit mutations, we analyzed the functions of predicted topological regions and structural domains. While these results agree with many previous predictions, we also discovered the unexpected function of several additional domains within the TFIIH core and kinase module subunits. Using site-specific cross-linking, we also mapped the interactions between a conserved Rad3 domain and the kinase module, with the results pointing to an unusual interaction interface.
MATERIALS AND METHODS
Genetic analysis.
Yeast plasmid shuffle strains listed in Table 1 were transformed with plasmids containing TFIIH subunit deletions, and transformants were selected on glucose complete medium lacking leucine (GC Leu−) at 30°C. To stabilize the association of Ssl2 with TFIIH and facilitate studies of subunit association, all strains except where noted contained a deletion in TFB6, a nonessential gene which promotes the reversible dissociation of Ssl2 from TFIIH (29). The Ssl2 mutants were also tested in a TFB6 wild-type (WT) strain (Table 1, SHY706). Three independent transformants for each derivative were patched on 5-fluoroorotic acid (5-FOA) at 30°C to select for cells that had lost the WT Ura+ plasmid and to test for viability of each deletion construct. Yeast strains that were 5-FOA resistant were tested for growth by streaking for single colonies on GC Leu− medium at 18, 25, 30, and 37°C. To test for UV sensitivity, 5-FOA-resistant yeast strains were grown overnight in yeast extract-peptone-dextrose (YPD) medium with adenine at 30°C. Cultures were diluted to an A600 of 0.7 and serially diluted in 10-fold increments. Three microliters of diluted cells was spotted on YPD-adenine plates, and plates were exposed to 0, 10, 20, or 40 mJ/m2 UV light (Stratalinker 1800; Stratagene) and then incubated at 30°C for 2 days.
TABLE 1.
S. cerevisiae strains used in this study
| Strain name | Function | Genotype |
|---|---|---|
| SHY904 | SSL1 shuffle strain | matα Δade2::hisG his3-Δ200 leu2-Δ0 lys2-Δ0 met15-Δ0 trp-Δ63 ura3-Δ0 ssl1Δ::HPH tfb6Δ::KanMX pSH1594(ARS CEN URA3 SSL1) |
| SHY873 | SSL2 shuffle strain (tfb6Δ) | matα Δade2::hisG his3-Δ200 leu2-Δ0 lys2-Δ0 met15-Δ0 trp1-Δ63 ura3-Δ0 ssl2Δ::HPH tfb6Δ::KanMX pSH620(ARS CEN URA3 SSL2) |
| SHY706 | SSL2 shuffle strain (TFB6) | matα Δade2::hisG his3-Δ200 leu2-Δ0 lys2-Δ0 met15-Δ0 trp1-Δ63 ura3-Δ0 ssl2Δ::KanMX pSH620(ARS CEN URA3 SSL2) |
| SHY906 | TFB2 shuffle strain (tfb6Δ) | matα Δade2::hisG his3-Δ200 leu2-Δ0 lys2-Δ0 met15-Δ0 trp1-Δ63 ura3-Δ0 tfb2Δ::HPH tfb6Δ::KanMX pSH1596(ARS CEN URA3 TFB2) |
| SHY907 | TFB3 shuffle strain (tfb6Δ) | matα Δade2::hisG his3-Δ200 leu2-Δ0 lys2-Δ0 met15-Δ0 trp1-Δ63 ura3-Δ0 tfb3Δ::HPH tfb6Δ::KanMX pSH1597(ARS CEN URA3 TFB3) |
| SHY908 | TFB4 shuffle strain (tfb6Δ) | matα Δade2::hisG his3-Δ200 leu2-Δ0 lys2-Δ0 met15-Δ0 trp1-Δ63 ura3-Δ0 tfb4Δ::HPH tfb6Δ::KanMX pSH1598(ARS CEN URA3 TFB4) |
| SHY872 | RAD3 shuffle strain (tfb6Δ) | matα Δade2::hisG his3-Δ200 leu2-Δ0 lys2-Δ0 met15-Δ0 trp1-Δ63 ura3-Δ0 tfb6Δ::HPH rad3Δ::KanMX pJF63(ARS CEN URA3 RAD3) |
Plasmids.
Deletion constructs of TFIIH subunits are derivatives of the LEU2-containing plasmids listed in Table 2. All analyzed genes contained a single copy of the Flag epitope and a tandem affinity purification (TAP) tag at the C terminus-coding end. Tfb3 and Tfb4 deletion constructs contain a glycine-serine linker (GSGSGS) in place of the deleted segment. Ssl1, Ssl2, and Tfb2 mutants do not contain a linker.
TABLE 2.
Plasmids used in this study
| Plasmid name | Description |
|---|---|
| pSH1539 | ARS CEN LEU2 SSL1-Flag1-TAP |
| pLH240 | ARS CEN LEU2 SSL2-Flag1-TAP |
| pSH1541 | ARS CEN LEU2 TFB2-Flag1-TAP |
| pSH1542 | ARS CEN LEU2 TFB3-Flag1-TAP |
| pSH1543 | ARS CEN LEU2 TFB4-Flag1-TAP |
| pSH1594 | ARS CEN URA3 SSL1 |
| pSH620 | ARS CEN URA3 SSL2 |
| pSH1596 | ARS CEN URA3 TFB2 |
| pSH1597 | ARS CEN URA3 TFB3 |
| pSH1598 | ARS CEN URA3 TFB4 |
| pJF63 | ARS CEN URA3 RAD3 |
| pSH1599 | 2μm LEU2 pADH1-RAD3-13 Myc |
TAP IPs, Western blot analysis, and quantitation.
TAP-tagged Ssl1, Ssl2, Tfb2, Tfb3, and Tfb4 (WT or deletion derivative) were immunoprecipitated from Saccharomyces cerevisiae whole-cell extracts with IgG-Sepharose Fast Flow beads (GE Healthcare Biosciences) essentially as described previously (31) except that the TAP whole-cell extract and immunoprecipitation (IP) buffer used was 50 mM HEPES (pH 7.6 at 4°C), 250 mM potassium acetate (KOAc), 1 mM magnesium acetate (MgOAc), 1 mM EDTA, 20% glycerol, and 1 mM dithiothreitol (DTT) with protease inhibitors as previously described. These strains contained both nontagged WT and TAP-tagged WT proteins or deletion derivatives. TFIIH was eluted from the beads by tobacco etch virus (TEV) protease cleavage and analyzed by Western blotting using polyclonal rabbit antibodies against Ssl2 (3079), Tfb2 (3141), Tfb4 (3113), Ssl1 (3167), Tfb3 (3088), and Rad3 (3109) (all, Hahn lab), Tfb5 (Ranish lab), and Kin28 (PRB-260C; Covance) or the monoclonal mouse antibody against calmodulin binding protein (Santa Cruz). Protein signals were quantified using Odyssey scanner software (Li-Cor) by generating a standard curve using a titration from a WT TAP-tagged IP. Each protein analyzed was normalized to the amount of the wild-type TAP-tagged protein obtained by IP. Reported results are from at least two independent experiments.
In vivo Rad3-BPA cross-linking.
A structure model was built for yeast Rad3 to identify probable surface-exposed residues in the Arch and FeS domains. Models were built using the S. cerevisiae Rad3-Archaeoglobus fulgidus XPD alignment of Fan et al. (47) and Modeler, version 9.9 (33). Top models were analyzed by Verify3D (34) and MolProbity (35). After the models were used to identify likely surface-exposed residues, 24 sites, most of which are not absolutely conserved in Rad3/XPD proteins, were chosen for insertion of p-benzoyl-l-phenylalanine (BPA). The chosen codons were replaced with amber stop codons in plasmid pSH1599 (Table 2) that contains the ADH1 promoter driving expression of Rad3 with a 13-Myc C-terminal tag on a 2μm plasmid expressing LEU2. Cells were cotransformed to a RAD3 shuffle strain SHY872 (Table 1) containing a plasmid expressing URA3 and RAD3 and chromosomal deletions of the RAD3 and TFB6 genes as well as plasmid pLH157 encoding a tRNA/BPA-tRNA synthetase which inserts BPA at amber codons (36, 37). Cells which had lost the RAD3 WT plasmid were selected on FOA plates containing 1 mM BPA.
For in vivo cross-linking (38), strains were grown overnight at 30°C in synthetic glucose complete medium lacking leucine and tryptophan with 1 mM BPA until the A600 was 1.5 to 2.6. Five milliliters of the culture was placed into the well of a six-well cell culture dish and irradiated with 365-nm UV light in a Spectrolinker XL-1500a UV cross-linker (Spectronics Corp.). Cells were irradiated three times at an energy setting of 8,500 × 100 μJ/cm2 with 2-min rests on ice between treatments. Cells were washed with 1 ml of H2O and frozen at −70°C. Cells were later thawed in 200 μl of 0.1 M NaOH, incubated for 5 min at room temperature, pelleted in a microcentrifuge, suspended in reducing SDS-PAGE buffer, and analyzed by Western blotting after electrophoresis in 3 to 8% acrylamide Tris-acetate NuPAGE gels (Invitrogen).
RESULTS
Experimental strategy for analysis of TFIIH function.
To systematically examine functions of the TFIIH subunits, we generated a series of internal deletions within three core subunits (Ssl1, Tfb4, and Tfb2), in the translocase subunit Ssl2, and in Tfb3, the subunit that links the kinase module to TFIIH. A similar approach has been applied to the core subunit Tfb1 that contains the Anchor region (30). Our mutations were designed to test the importance of predicted topological regions and domains within the TFIIH subunits. All the mutated subunits contained one copy of the Flag and TAP tag epitopes at the C terminus. The phenotypes of the mutations were first analyzed by yeast plasmid shuffle assay, where the mutant copy of the gene was substituted for the wild type. To enhance Ssl2 stability within TFIIH for subsequent IP analysis, the plasmid shuffle strains, except where noted, contained a deletion in TFB6, the nonessential factor that facilitates dissociation of Ssl2 from TFIIH (29). The nonlethal TFIIH subunit mutations were tested for growth of single colonies at 18 to 37° and for UV sensitivity as a measure of NER function. So that both lethal and nonlethal mutations could be analyzed for defects in TFIIH subunit association, cells containing both the TAP-tagged mutant subunit and nontagged wild-type subunit were used in IP assays. Whole-cell extracts from these strains were used to immunoprecipitate the mutant subunit via the TAP tag. Association of the mutant subunit with other TFIIH subunits was analyzed and quantitated by Western blotting in at least two independent experiments.
Functional regions of the Ssl1-Tfb4 heterodimer.
Although divergent in sequence, the Tfb4 and Ssl1 subunits have similar domain organizations with N-terminal von Willebrand factor A (VWA) domains and C-terminal Ring fingers (39, 40) (Fig. 2 and 3). Based on cross-linking and other information, we proposed that these two subunits dimerize to form the base of the TFIIH core (Fig. 1) (30). From previous studies, both VWA domains are proposed to interact with each other's VWA and Ring domains. Additionally, the Tfb4 and Ssl1 VWA domains show conserved cross-links with the Anchor region of Tfb1 and the Hub region of Tfb2 (30). The Tfb4 VWA also cross-links to the Tfb2 HEAT domain. Finally, Ssl1 has a unique 116-residue N-terminal extension (NTE) that cross-links to the Tfb3 Latch domain, the Ssl2 Lock-N region, and the Rad3 helicase domain 2 (HD2). In sum, there are predicted interactions between the proposed Ssl1-Tfb4 heterodimer and most other TFIIH subunits.
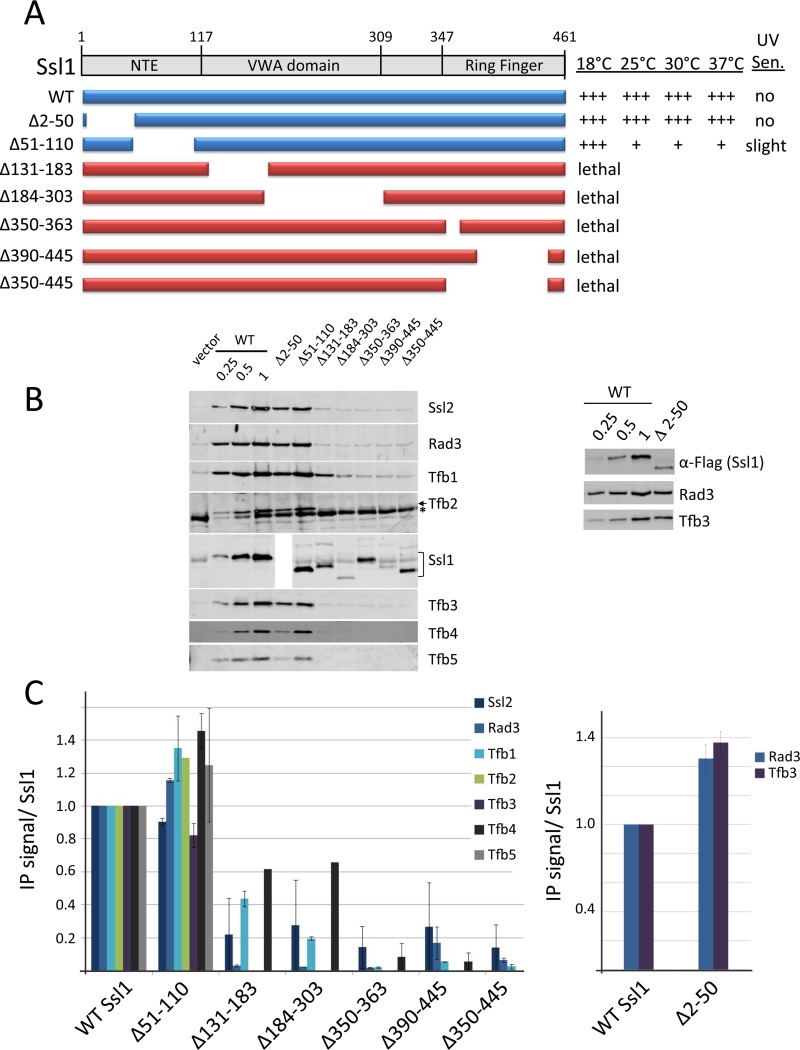
FIG 2.
Analysis of Ssl1 functional regions. (A) Schematic of Ssl1 domains and deletion derivatives. Lethal mutations are shown in red, and viable mutations are in blue. The relative growth of these derivatives at various temperatures and UV sensitivity at 30°C are indicated. (B) Whole-cell extracts from strains containing untagged WT Ssl1 and TAP-tagged Ssl1 derivatives were subjected to IP and Western analysis using the indicated TFIIH antisera (left). Variable amounts of the IP from cells with WT TAP-tagged Ssl1 were loaded to allow quantitation of the signals from the mutant derivatives. An asterisk indicates a nonspecific Western signal. The polyclonal Ssl1 antiserum reacts poorly with the Ssl1 Δ2–50 mutant. A parallel IP was done with this mutant probing for the Ssl1 Flag tag (right panel). (C) Quantitation of Western analysis from two independent experiments. Error bars indicate standard errors of the means.
FIG 3.
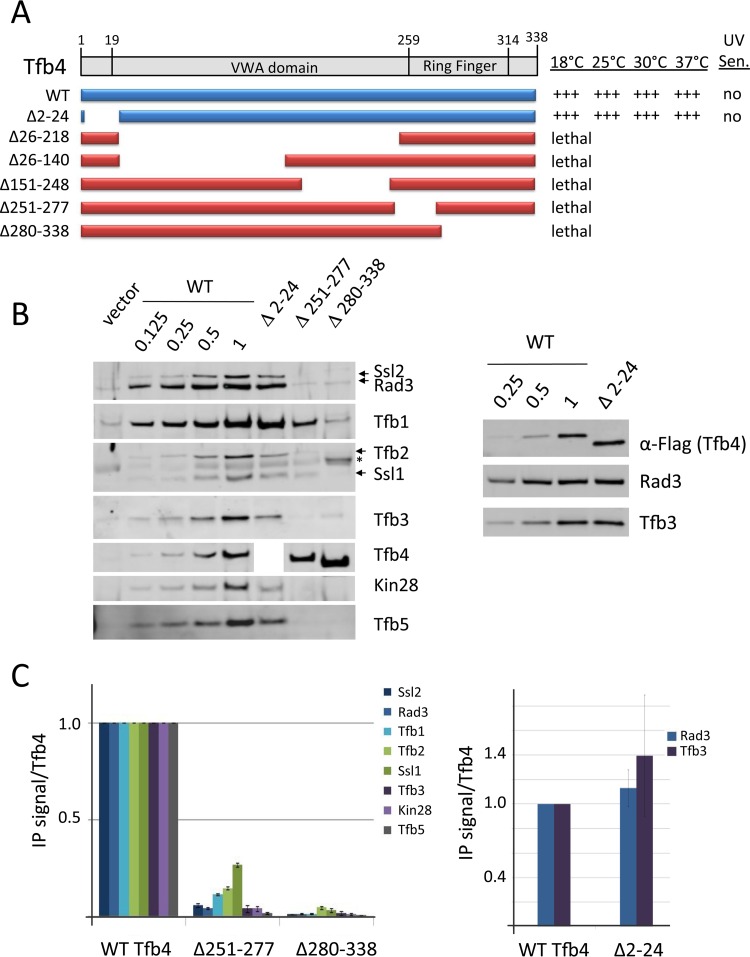
Analysis of Tfb4 functional regions. (A) Schematic of Tfb4 domains and deletion derivatives. Lethal mutations are shown in red, and viable mutations are in blue. The relative growth of these derivatives at various temperatures and UV sensitivity at 30°C are indicated. (B and C) Western analysis of IP reactions and quantitation as described in the legend to Fig. 2. An asterisk indicates a nonspecific Western signal. The polyclonal Tfb4 antiserum reacted poorly with the Tfb4 Δ2–24 mutant. A parallel IP was done with this mutant probing for the Tfb4 Flag tag (right panel). Error bars indicate standard errors of the means.
The Ssl1 NTE was targeted by two deletions, of residues 2 to 50 (Ssl1 Δ2–50) and 51 to 110 (Ssl1 Δ51–110) (Fig. 2A). Ssl1 containing a partial deletion of the NTE (Ssl1 Δ2–50) coprecipitated with all tested TFIIH subunits (Fig. 2B, left). Our Ssl1 polyclonal antiserum did not react well with this derivative. However, probing a parallel IP experiment with anti-Flag antiserum showed a nearly normal ratio of coprecipitating Rad3 and Tfb3 with Ssl1 Δ2–50, despite the fact that protein levels of this Ssl1 derivative are two to three times lower than the WT Ssl1 level (Fig. 2B, right). We conclude that this mutant does not have a defect in TFIIH subunit association. Combined with the result that this deletion has no growth or UV-sensitive phenotype (Fig. 2A), this suggests that Ssl1 residues 2 to 50 are not important for TFIIH function.
Strains with deletion of Ssl1 residues 51 to 110 (Ssl1 Δ51–110) grew very slowly at all temperatures tested except 18°C and had a slight UV-sensitive phenotype (Fig. 2A). However, co-IP of other TFIIH subunits with Ssl1 Δ51–110 was nearly normal (Fig. 2B and C). Given the numerous cross-links observed between this region and other TFIIH subunits, it was surprising that TFIIH subunit association was unchanged. Since this mutant has a strong growth phenotype, we speculate that it may perturb one of the TFIIH enzymatic functions. For example, because the Ssl1 NTE cross-links to the Ssl2 Lock-N region, one possibility is that Ssl1 Δ51–110 is defective in a regulatory interaction between Ssl1 and Ssl2, analogous to the regulatory interaction between the Tfb2-Tfb5 heterodimer and the Ssl2 Lock domain (30, 41).
In contrast to the two N-terminal mutations, deletion constructs with portions of the Ssl1 VWA domain removed were defective for all tested TFIIH subunit interactions. Surprisingly, the Ssl1 Ring finger is also essential for Ssl1 interaction with all TFIIH subunits even though no conserved cross-links between the Ring and other TFIIH subunits have been observed (30).
We found that the Tfb4 VWA domain is essential for protein stability as two mutations that removed large segments of VWA eliminated detectable Tfb4 (data not shown). Deletion of the Tfb4 Ring domain showed that it is surprisingly essential for association with other TFIIH subunits, the same phenotype observed upon deletion of the Ssl1 Ring domain (Fig. 3). The Tfb4 Ring did not show conserved cross-links with any TFIIH subunit outside Ssl1 (30). As expected, none of the Ssl1 or Tfb4 VWA or Ring finger mutations supported yeast growth. Tfb4 with a deletion of the N-terminal extension (residues 2 to 24) coprecipitated with all tested TFIIH subunits. Our Tfb4 polyclonal antiserum did not react well with this derivative. However, probing a parallel IP experiment with anti-Flag antiserum showed nearly normal ratios of coprecipitating Rad3 and Tfb3. We conclude that this mutant does not have a defect in TFIIH subunit association.
Tfb2 and the Hub domain.
Tfb2 contains a predicted N-terminal HEAT repeat domain and a C-terminal region termed the Hub (Fig. 4A). The Hub binds Tfb5/p8, and this complex modulates Ssl2 activity. This interaction was also proposed to anchor Ssl2/XPB to TFIIH (30, 41). The Hub region shows conserved cross-links with the VWA domains of Ssl1 and Tfb4 (30). In contrast, no conserved cross-links have been observed between the HEAT domain and other subunits of TFIIH.
FIG 4.
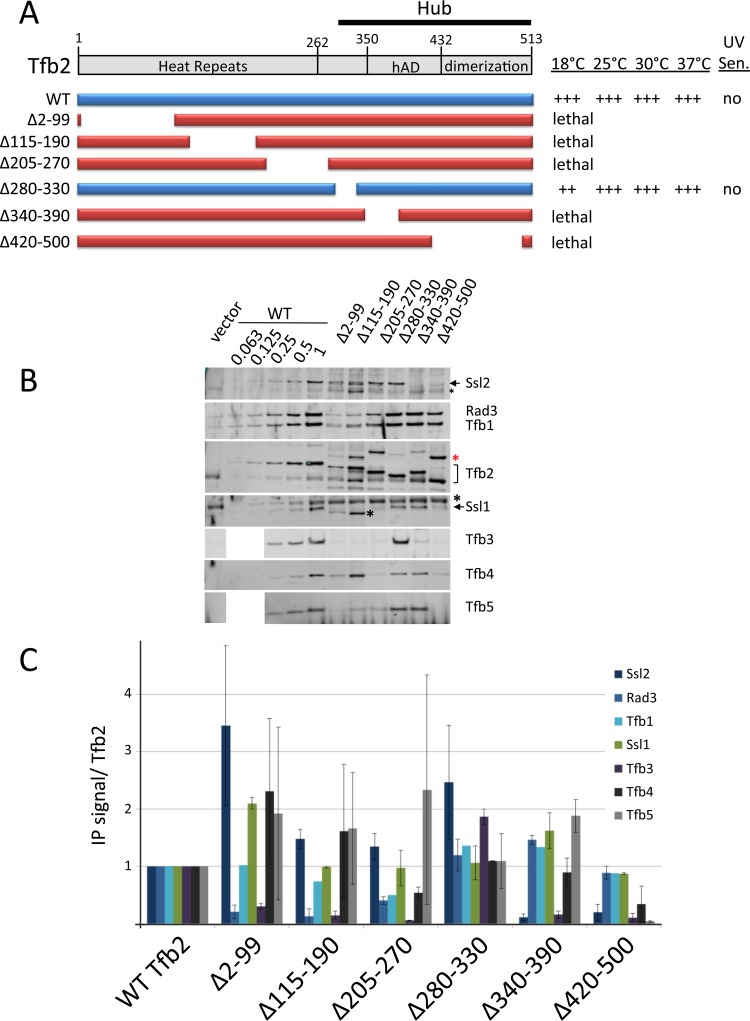
Analysis of Tfb2 functional regions. (A) Schematic of Tfb2 domains and deletion derivatives. Lethal mutations are shown in red, and viable mutations are in blue. The relative growth of these derivatives at various temperatures and UV sensitivity at 30°C are indicated. (B and C) Western analysis of IP reactions and quantitation as described in the legend to Fig. 2. A black asterisk indicates a nonspecific Western signal, and a red asterisk indicates a fraction of Tfb2 not cleaved by TEV protease. Error bars indicate standard errors of the means.
As predicted, deletions within the Tfb2 Hub led to defects in association with Ssl2, Tfb5, and Tfb4 (Fig. 4). Tfb2 Δ340–390, with the predicted helical domain (Fig. 4, hAD) at the Hub N terminus removed, is defective in Ssl2 association. Tfb2 Δ420–500, with a deletion of the Hub C terminus, is defective for association with Ssl2 as well as Tfb4 and Tfb5. Both Hub deletion constructs were also defective in Tfb3 association, a result not expected from protein cross-linking assays. Also surprising was that all three HEAT domain deletion constructs (Tfb2 Δ2–99, Δ115–190, and Δ205–270) were defective in Tfb3 association, a result not predicted from previous work. Finally, a mutant with a deletion at the C terminus of the HEAT domain (residues 205 to 270) is partially defective for Tfb4 association. This is consistent with nonconserved cross-links observed between this region of Tfb2 and the VWA domain of Tfb4 (30). In sum, the Tfb2 Hub, as predicted, is required for stable interaction with Ssl2, Tfb3, Tfb4, and Tfb5 while the HEAT domain has an unexpected role in association with Rad3, Tfb3, and Tfb4. Also surprising is the finding that every lethal mutation in Tfb2 abrogated interaction with Tfb3. It is possible that the loss of Tfb3 in these mutations is due to altered interactions between Ssl2 and the Tfb2 derivatives.
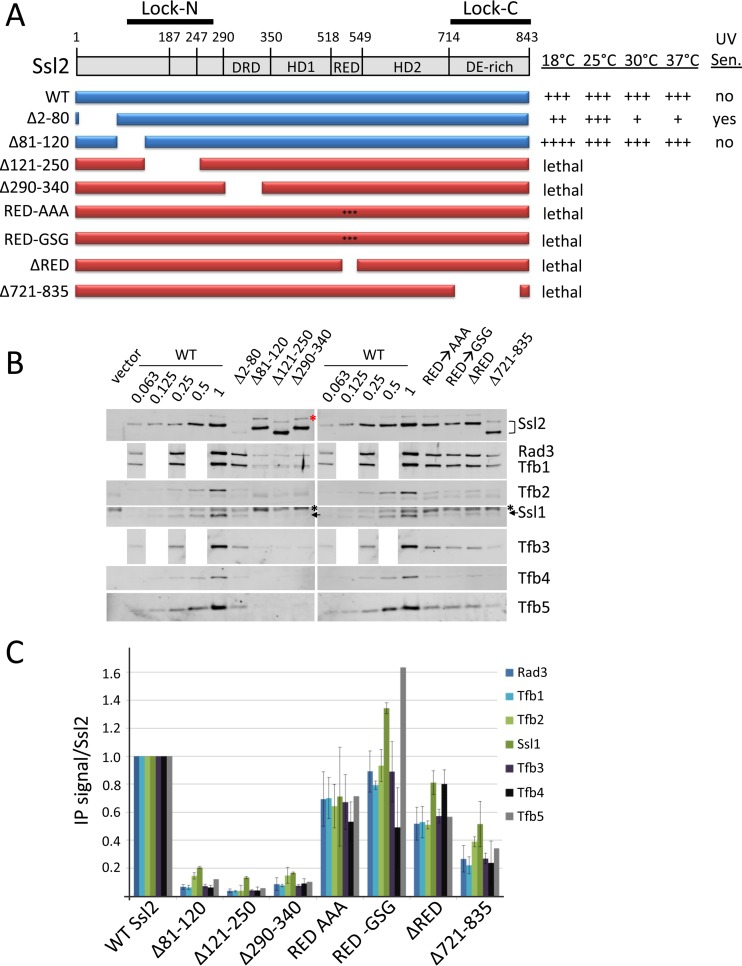
Ssl2 and the Lock domain.
Eukaryotic Ssl2/XPB contains N- and C-terminal extensions (termed the N- and C-terminal Lock regions), and the remainder of Ssl2/XPB domain architecture is conserved in archaea (30, 32) (Fig. 5). In archaea, XPB plays a role in NER, and there is no known TFIIH-like complex. Conserved intra- and intermolecular cross-links and other data suggested that the N- and C-terminal Lock extensions associate to form the Lock region, a domain proposed to be involved in interactions with the Tfb2-Tfb5 complex, Ssl1, Tfb3, and Rad3 HD2 (30). This region also interacts with the NER exonuclease XPG (42).
FIG 5.
Analysis of Ssl2 functional regions. (A) Schematic of Ssl2 domains and deletion derivatives. The RED motif was either deleted or replaced by the residues GSG or AAA, as shown. The relative growth of these derivatives at various temperatures and UV sensitivity at 30°C are indicated. (B and C) Western analysis of IP reactions and quantitation as described in the legend to Fig. 2. A black asterisk indicates a nonspecific Western signal, and a red asterisk indicates a fraction of Ssl2 deletion derivatives not cleaved by TEV protease. Error bars indicate standard errors of the means.
A strain containing a deletion of the N-terminal segment of Lock-N (residues 81 to 120) grew normally (except for faster growth at 18°C) and was not UV sensitive (Fig. 5). However, IP analysis showed that it had defective interactions with all tested TFIIH subunits. This result shows that the IP assays can be a more stringent test for defects in TFIIH subunit association than growth assays. As expected, deletion of the Lock-N residues 121 to 250 and the adjacent DRD domain (residues 290 to 340) was lethal, and this derivative showed severely reduced interactions with all other TFIIH subunits. Archaea and eukaryotic XPBs contain the conserved sequence motif R-E-D at the tip of a conserved loop that lies between HD1 and HD2, is unique to the XPBs, and is essential for helicase function (32). These three residues were essential for Ssl2 function as substitutions or deletion of the RED motif was lethal for growth, but this defect had only a modest effect on TFIIH subunit association with Ssl2. Finally, the deletion of the Lock-C region was lethal and reduced interaction of Ssl2 with all tested TFIIH subunits although this defect was not as severe as in the Lock-N mutations. In sum, our results confirm the importance of the Lock region and the adjacent DRD domain for TFIIH-Ssl2 association. We also confirm that the RED motif plays an essential role in Ssl2 function.
Unexpectedly, Ssl2 Δ2–80, with a deletion just N-terminal to the Lock-N region, gave a temperature- and cold-sensitive phenotype (Fig. 5A). However, IP analysis showed 5- to 10-fold lower levels of this derivative than of the WT (Fig. 5B), likely explaining the growth and UV-sensitive phenotypes. Since Tfb6 directly interacts with Ssl2 and stimulates dissociation from TFIIH, we tested all Ssl2 derivatives in a TFB6 WT strain. The only difference observed was in the Ssl2 Δ2–80 derivative, which showed less defective growth and UV phenotypes. The TFB6 Ssl2 Δ2–80 strain showed only slight UV sensitivity, slow growth at 37°C, and slightly slower growth at 18°C.
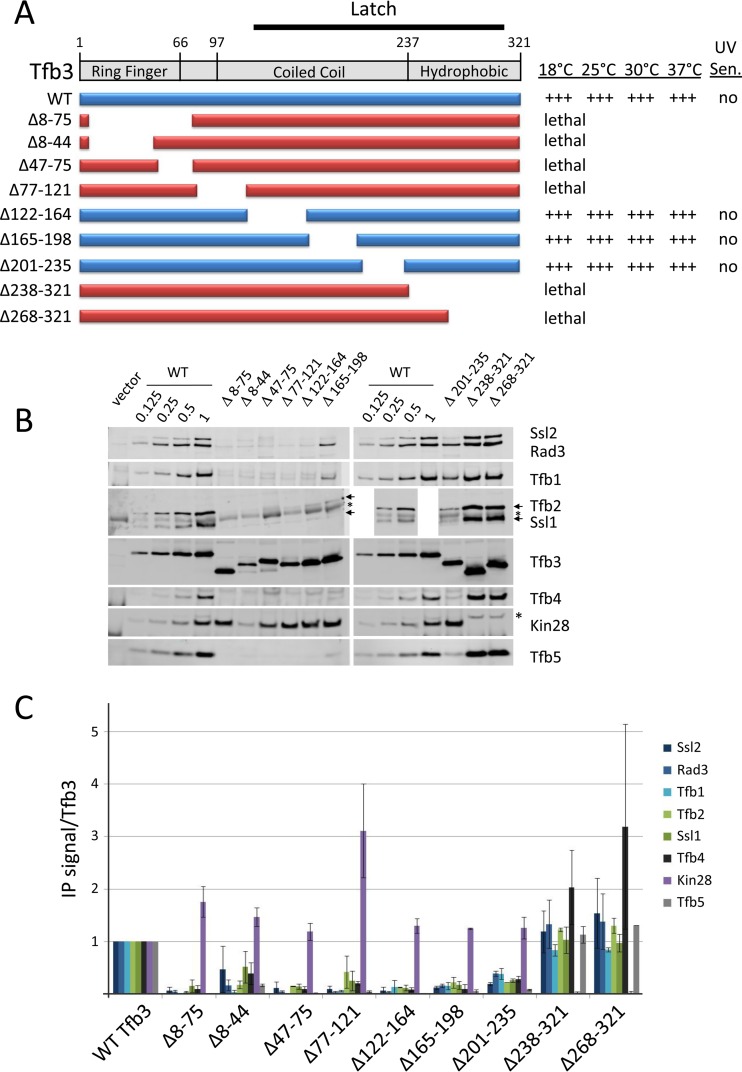
Tfb3 and the Latch region.
Tfb3, a subunit of the TFIIH kinase module, consists of an N-terminal Ring finger, a central coiled-coil domain, and a C-terminal hydrophobic region (Fig. 6A). The coiled-coil and hydrophobic regions were together proposed to form the Latch region, functioning to anchor the kinase module to Rad3, Ssl2, and Ssl1 (30). The Tfb3 hydrophobic region is known to interact with the other two kinase module subunits, Kin28/CDK7 and Ccl1/cyclin H (43). We found that deletions of the Tfb3 Ring finger domain are lethal and surprisingly disrupt interactions with all other tested TFIIH subunits except Kin28 (Fig. 6). However, yeast-specific cross-links were detected between the Tfb3 Ring finger and Rad3 and Tfb1 (30), consistent with our new results. Deletions covering most of the coiled-coil domain are surprisingly viable. However, IP analysis shows that most of these mutants are defective in interactions with all subunits except Kin28 (Fig. 6). Finally, mutants with deletions of the C-terminal hydrophobic domain (residues 238 to 321 and 268 to 321) lose interaction with Kin28, as predicted from previous work (43). None of the nonlethal strains showed a UV-sensitive phenotype. In sum, our results point to an unexpected role for the Tfb3 Ring finger domain in TFIIH subunit association and confirm the predicted roles for the Ring finger in TFIIH function (43) and for the Latch domain in Tfb3-TFIIH association.
FIG 6.
Analysis of Tfb3 functional regions. (A) Schematic of Tfb3 domains and deletion derivatives. The relative growth of these derivatives at various temperatures and UV sensitivity at 30°C are indicated. (B and C) Western analysis of IP reactions and quantitation as described in the legend to Fig. 2. An asterisk indicates a nonspecific Western signal. Error bars indicate standard errors of the means.
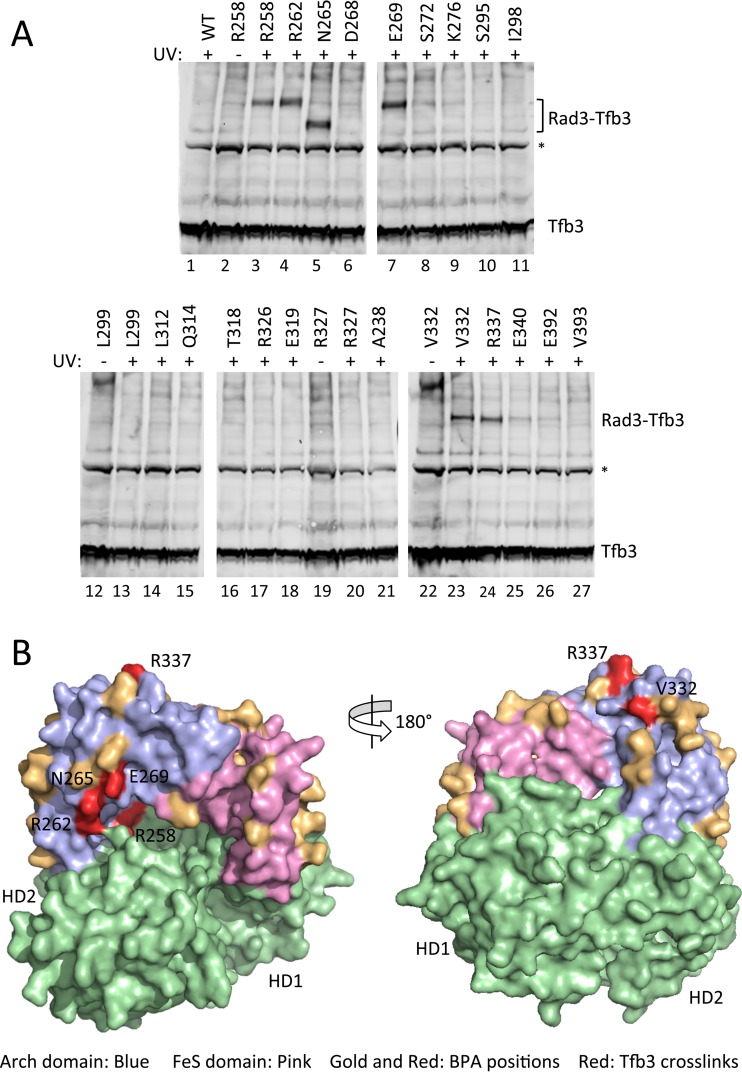
Mapping the TFB3-Rad3 Arch domain interaction.
Rad3/XPD contains two structured regions, termed the FeS and Arch domains, inserted within HD1 (44–47) (Fig. 7). The iron sulfur cluster domain (FeS) plays an important role in Rad3 helicase function as many mutations in this domain decrease helicase activity (9, 10, 44). These FeS mutations are associated with XP, a disease linked to defects in NER. First recognized in archaeal XPD, the Arch domain, together with the FeS domain, forms an enclosed tunnel that was proposed to be involved in single-stranded DNA binding (47). At least two human Arch domain mutations were shown to cause trichothiodystrophy (TTD), the most severe form of TFIIH-associated disease. Protein-protein interaction studies have implicated the Arch domain as interacting with Tfb3/Mat1 and the Mediator subunit Med11 (9, 26, 30).
FIG 7.
Mapping the Rad3 Arch domain-Tfb3 interface. (A) Strains containing the photoreactive nonnatural amino acid BPA inserted in the Rad3 Arch domain were subjected to UV. Whole-cell extracts were made from these strains and analyzed by Western blotting, probing for Tfb3. Tfb3 and the Rad3-Tfb3 cross-linking products are indicated. A nonspecific band is indicated by an asterisk. (B) Structure model for Rad3 indicating positions of BPA insertions and Tfb3 cross-links. The Rad3 Arch domain is shown in blue, the Rad3 FeS domain is in pink, and the two Rad3 helicase domains are in green. Positions of BPA insertions which did not cross-link in the Arch and FeS domains are shown in gold, and positions that strongly cross-linked to Tfb3 are in red.
To map in vivo interactions with both of these domains, we inserted the nonnatural photoreactive amino acid BPA at specific positions within the Arch and FeS (Table 3). To accomplish this, amber nonsense mutations were substituted for selected codons within Rad3, and these mutations were suppressed by the presence of a plasmid containing an archaeal amber suppressor tRNA and a tRNA synthetase that specifically charges this tRNA with BPA (36, 37) (see Materials and Methods). Surface-exposed positions on the two domains were predicted using structure models for Rad3, based on the structure of archaeal XPD (45, 47). Most residues targeted for BPA insertion were selected because they were near conserved residues. After suppression of the nonsense mutations with BPA, all derivatives expressed Rad3 at levels approximately the same as a wild-type strain.
TABLE 3.
Positions of BPA insertions in the Rad3 Arch and FeS domains
| Domain and BPA insertion position | Tfb3 cross-linka |
|---|---|
| Arch | |
| R258 | + |
| R262 | + |
| N265 | + |
| D268 | − |
| E269 | + |
| S272 | − |
| K276 | − |
| S295 | − |
| I298 | − |
| L299 | − |
| L312 | − |
| Q314 | − |
| T318 | − |
| R326 | − |
| E319 | − |
| R327 | − |
| A328 | − |
| V332 | + |
| R337 | + |
| E340 | Weak |
| E392 | Weak |
| V393 | − |
| FeS | |
| V120 | − |
| R134 | − |
| L155 | − |
| E157 | − |
| Y163 | − |
| N164 | − |
| E166 | − |
| E168 | − |
| E179 | − |
| E186 | − |
| L202 | − |
Cross-linking is indicated as follows: +, strong; −, not detectable; weak, detectable but weak.
Twenty-two Rad3 derivatives with BPA in the Arch domain were tested for in vivo cross-linking. Five milliliters of cells grown in synthetic BPA-containing medium was UV cross-linked, and total protein was extracted and analyzed by Western blotting (38). When probing for Tfb3, a known TFIIH subunit that interacts with the Arch domain, we found that six Rad3-BPA derivatives cross-linked strongly to Tfb3 (R258, R262, N265, E269, V332, and R337) while two other derivatives (E340 and E392) showed weak but detectable cross-linking (Fig. 7A). Cross-linked Rad3 N265 BPA (Fig. 7, lane 5) showed faster migration than other derivatives as this construct has lost the Rad3 C-terminal Myc tag. In contrast, probing for Myc-tagged Rad3 in some cases showed weakly visible higher-molecular-weight UV-dependent products, but none of these correspond to the size of the Rad3-Tfb3 cross-links (data not shown). Evidently, only a small fraction of Rad3 cross-links to Tfb3. This suggests that Tfb3 might be substoichiometric in the TFIIH complex in vivo or that a significant percentage of Rad3 is not incorporated into TFIIH. Mapping the Tfb3 cross-linked positions on the Rad3 structure model (Fig. 7B, red residues) shows two clusters of cross-links, separated on either side of the Arch domain (blue). This suggests that Tfb3 interacts with the Arch domain on opposite faces, possibly using two distinct regions of Tfb3. BPA positions which did not show detectable cross-links are shown in gold. We also probed this blot with available antisera directed against Ssl1, Tfb1, Med6, Med17, and Med22 (Med 11 antiserum is not available), but no cross-links were observed. BPA insertions were similarly targeted to 11 positions on the predicted surface of the FeS domain and subjected to in vivo UV cross-linking. None of these derivatives showed detectable cross-links (Table 3).
DISCUSSION
TFIIH is the most complex RNA Pol II basal transcription factor and plays essential roles in transcription and NER. Previous studies have mapped intermolecular interactions among TFIIH subunits and identified mutations in three subunits causing human disease. We recently proposed a model for the architecture of TFIIH based on conserved protein-protein cross-links and structural, biochemical, and genetic data. Here, we used these predictions as a guide to test the role of the predicted domains and conserved topological regions within TFIIH. While our results generally agree with the predicted intermolecular subunit interactions, we find important roles for additional domains within the TFIIH subunits that were not predicted from previous studies.
The Ssl1-Tfb4 heterodimer.
Our earlier work predicted that a heterodimer of Ssl1 and Tfb4 formed the base of the TFIIH core domain. The subunits have similar domain architectures and have numerous intermolecular cross-links with each other and with many other TFIIH core subunits. Our first surprise was finding that deletion of residues 51 to 110 in the Ssl1 NTE domain gave rise to a severe growth phenotype and slight UV sensitivity while not affecting TFIIH subunit association. Since the Ssl1 NTE cross-links to the Ssl2/XPB Lock region, one possible explanation for the observed phenotype is that the NTE modulates Ssl2 enzymatic function in a manner similar to Tfb5/Tfb1, which also interacts with the Ssl2/XPB Lock region. Ssl1 residues 76 to 93 are modestly conserved with human p44 although, as yet, no disease-causing mutations have been found in this subunit.
As expected, deletions within the VWA domains of Ssl1 and Tfb4 were lethal and caused severe defects in TFIIH subunit association and/or protein stability. Deletions within both the Ssl1 and Tfb4 Ring domains were lethal and caused severe defects in TFIIH subunit association. While the Ring domains showed no cross-links to other TFIIH subunits that are conserved between yeast and humans, the Ssl1 Ring cross-links to Tfb4, Tfb2, and Tfb1 while the Tfb4 Ring cross-links to Ssl1 and Tfb1. Two possibilities are (i) that deletion of the Ring domains indirectly affects TFIIH subunit association by disrupting the conformation of the VWA domains or (ii) that the Ring domains make essential interactions with other TFIIH subunits such as Tfb2 and Tfb1.
Tfb2 and the Hub region.
Previous studies showed that the Tfb2/p52 Hub domain binds Tfb5/p8 and that this heterodimer binds the Ssl2/XPB Lock domain to modulate Ssl2 function. Consistent with this, deletion of the Hub N-terminal region (residues 340 to 390) is lethal and causes a defect in Ssl2 interaction while deletion of the Hub C-terminal region (residues 420 to 500) causes a defect in both Ssl2 and Tfb5 binding. Deletions within the Tfb2 HEAT repeat domain were lethal, and the mutants were surprisingly defective for interactions with Rad3 and Tfb3. Only one cross-link was previously observed between human p52 and HD1 of XPD, and none was observed between Tfb2/p52 and Tfb3. Although no direct interactions between Tfb2 and Tfb3 have been detected before, the two proteins seem tightly linked since five Tfb2 mutations covering nearly all of the Tfb2 protein caused defects in Tfb3 interaction. Our mutations have confirmed the role of the Hub domain and revealed an unexpected function of the HEAT repeats and a link between Tfb2 and Tfb3.
Ssl2 Lock region and the RED motif.
As described above, Ssl2 function is modulated through interactions with Tfb2/Tfb5. Deletion of both Ssl2 subdomains, Lock-N and Lock-C, were lethal, and these mutations disrupted interactions with all tested TFIIH subunits. This finding is in agreement with the model that Tfb2/Tfb5 anchors Ssl2 to TFIIH. The conserved sequence motif RED, found within a loop between Ssl2 HD1 and HD2, was proposed to be essential for Ssl2 function, and a mutation in this motif was previously found to impair human NER (5). Both deletion and substitution of this motif were lethal but had no effect on TFIIH subunit association, supporting the model that this motif is required for DNA translocase function.
Tfb3 and the Latch region.
Tfb3 contains an N-terminal Ring finger and a C-terminal segment termed the Latch region. The Latch cross-links to multiple domains of Ssl2 and Rad3 as well as to Ssl1, and the Latch is proposed to anchor the kinase module to TFIIH. Given the numerous cross-links between the Latch and other TFIIH subunits, it was surprising that none of the Latch deletions resulted in a growth or UV-sensitive phenotype. However, our IP analysis showed that these mutations caused defects in association with all other TFIIH subunits tested except the kinase module subunit Kin28. As predicted from previous work, kinase binding was specifically abrogated by deletion of the hydrophobic C terminus of the Latch. In contrast to the nonessential function of the Latch coiled-coil region, deletion of the Tfb3 Ring domain was lethal and also eliminated all TFIIH subunit binding except for that of Kin28. The Ring domain showed no conserved cross-links with other TFIIH subunits although the yeast Tfb3 Ring cross-links to both Tfb1 and Rad3. Our results suggest that the Ring domain plays an essential role in stabilizing the association of Tfb3 with core TFIIH, consistent with previous results showing that the Ring domain is important for TFIIH transcription function (43).
Rad3 Arch-Tfb3 binding.
Site-specific in vivo cross-linking mapped the interaction interface between Rad3 and Tfb3. By positioning the photoreactive residue BPA on the predicted surface of the Rad3 Arch, a domain previously shown to bind Tfb3, we localized the site of Tfb3 interaction. We found an unexpected Rad3-Tfb3 interface with Tfb3 cross-linking to opposite faces of the Arch. This suggests that more than one domain of Tfb3 binds the Arch. Using an analogous strategy, we were unable to detect factors binding to the Rad3 FeS domain, known to be essential for Rad3 function.
Our combined results have confirmed previous models for TFIIH subunit association and revealed unexpected roles for several of the subdomains within TFIIH. Our results will be important for guiding future studies on the function and assembly of TFIIH and its association with other transcription and NER factors.
ACKNOWLEDGMENTS
We thank members of the Hahn laboratory for advice and suggestions throughout the course of this work and Ivanka Kamenova for advice on in vivo cross-linking.
This work was supported by grant 2RO1GM053451 to S.H. and by P50 GM076547 and R21CA175849 to J.R.
REFERENCES
- 1.Compe E, Egly J-M. 2012. TFIIH: when transcription met DNA repair. Nat Rev Mol Cell Biol 13:343–354. doi: 10.1038/nrm3350. [DOI] [PubMed] [Google Scholar]
- 2.Grünberg S, Hahn S. 2013. Structural insights into transcription initiation by RNA polymerase II. Trends Biochem Sci 38:603–611. doi: 10.1016/j.tibs.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sainsbury S, Bernecky C, Cramer P. 2015. Structural basis of transcription initiation by RNA polymerase II. Nat Rev Mol Cell Biol 16:129–143. doi: 10.1038/nrm3952. [DOI] [PubMed] [Google Scholar]
- 4.Buratowski S, Hahn S, Guarente L, Sharp PA. 1989. Five intermediate complexes in transcription initiation by RNA polymerase II. Cell 56:549–561. doi: 10.1016/0092-8674(89)90578-3. [DOI] [PubMed] [Google Scholar]
- 5.Fuss JO, Tainer JA. 2011. XPB and XPD helicases in TFIIH orchestrate DNA duplex opening and damage verification to coordinate repair with transcription and cell cycle via CAK kinase. DNA Repair 10:697–713. doi: 10.1016/j.dnarep.2011.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Egly J-M, Coin F. 2011. A history of TFIIH: two decades of molecular biology on a pivotal transcription/repair factor. DNA Repair 10:714–721. doi: 10.1016/j.dnarep.2011.04.021. [DOI] [PubMed] [Google Scholar]
- 7.Winkler GS, Araújo SJ, Fiedler U, Vermeulen W, Coin F, Egly JM, Hoeijmakers JH, Wood RD, Timmers HT, Weeda G. 2000. TFIIH with inactive XPD helicase functions in transcription initiation but is defective in DNA repair. J Biol Chem 275:4258–4266. doi: 10.1074/jbc.275.6.4258. [DOI] [PubMed] [Google Scholar]
- 8.Tirode F, Busso D, Coin F, Egly JM. 1999. Reconstitution of the transcription factor TFIIH: assignment of functions for the three enzymatic subunits, XPB, XPD, and cdk7. Mol Cell 3:87–95. doi: 10.1016/S1097-2765(00)80177-X. [DOI] [PubMed] [Google Scholar]
- 9.Abdulrahman W, Iltis I, Radu L, Braun C, Maglott-Roth A, Giraudon C, Egly J-M, Poterszman A. 2013. ARCH domain of XPD, an anchoring platform for CAK that conditions TFIIH DNA repair and transcription activities. Proc Natl Acad Sci U S A 110:E633–42. doi: 10.1073/pnas.1213981110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuper J, Braun C, Elias A, Michels G, Sauer F, Schmitt DR, Poterszman A, Egly J-M, Kisker C. 2014. In TFIIH, XPD helicase is exclusively devoted to DNA repair. PLoS Biol 12:e1001954. doi: 10.1371/journal.pbio.1001954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fishburn J, Tomko E, Galburt E, Hahn S. 2015. Double-stranded DNA translocase activity of transcription factor TFIIH and the mechanism of RNA polymerase II open complex formation. Proc Natl Acad Sci U S A 112:3961–3966. doi: 10.1073/pnas.1417709112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grünberg S, Warfield L, Hahn S. 2012. Architecture of the RNA polymerase II preinitiation complex and mechanism of ATP-dependent promoter opening. Nat Struct Mol Biol 19:788–796. doi: 10.1038/nsmb.2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hahn S, Young ET. 2011. Transcriptional regulation in Saccharomyces cerevisiae: transcription factor regulation and function, mechanisms of initiation, and roles of activators and coactivators. Genetics 189:705–736. doi: 10.1534/genetics.111.127019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fazal FM, Meng CA, Murakami K, Kornberg RD, Block SM. 2015. Real-time observation of the initiation of RNA polymerase II transcription. Nature 525:274–277. doi: 10.1038/nature14882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kugel JF, Goodrich JA. 1998. Promoter escape limits the rate of RNA polymerase II transcription and is enhanced by TFIIE, TFIIH, and ATP on negatively supercoiled DNA. Proc Natl Acad Sci U S A 95:9232–9237. doi: 10.1073/pnas.95.16.9232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dvir A, Conaway RC, Conaway JW. 1997. A role for TFIIH in controlling the activity of early RNA polymerase II elongation complexes. Proc Natl Acad Sci U S A 94:9006–9010. doi: 10.1073/pnas.94.17.9006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lolli G, Johnson LN. 2005. CAK-cyclin-dependent activating kinase: a key kinase in cell cycle control and a target for drugs? Cell Cycle 4:572–577. [PubMed] [Google Scholar]
- 18.Coin F, Oksenych V, Mocquet V, Groh S, Blattner C, Egly J-M. 2008. Nucleotide excision repair driven by the dissociation of CAK from TFIIH. Mol Cell 31:9–20. doi: 10.1016/j.molcel.2008.04.024. [DOI] [PubMed] [Google Scholar]
- 19.Chang WH, Kornberg RD. 2000. Electron crystal structure of the transcription factor and DNA repair complex, core TFIIH. Cell 102:609–613. doi: 10.1016/S0092-8674(00)00083-0. [DOI] [PubMed] [Google Scholar]
- 20.Schultz P, Fribourg S, Poterszman A, Mallouh V, Moras D, Egly JM. 2000. Molecular structure of human TFIIH. Cell 102:599–607. doi: 10.1016/S0092-8674(00)00082-9. [DOI] [PubMed] [Google Scholar]
- 21.Gibbons BJ, Brignole EJ, Azubel M, Murakami K, Voss NR, Bushnell DA, Asturias FJ, Kornberg RD. 2012. Subunit architecture of general transcription factor TFIIH. Proc Natl Acad Sci U S A 109:1949–1954. doi: 10.1073/pnas.1105266109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He Y, Fang J, Taatjes DJ, Nogales E. 2013. Structural visualization of key steps in human transcription initiation. Nature 495:481–486. doi: 10.1038/nature11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murakami K, Tsai K-L, Kalisman N, Bushnell DA, Asturias FJ, Kornberg RD. 2015. Structure of an RNA polymerase II preinitiation complex. Proc Natl Acad Sci U S A 112:13543–13548. doi: 10.1073/pnas.1518255112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim TK, Ebright RH, Reinberg D. 2000. Mechanism of ATP-dependent promoter melting by transcription factor IIH. Science 288:1418–1422. doi: 10.1126/science.288.5470.1418. [DOI] [PubMed] [Google Scholar]
- 25.Miller G, Hahn S. 2006. A DNA-tethered cleavage probe reveals the path for promoter DNA in the yeast preinitiation complex. Nat Struct Mol Biol 13:603–610. doi: 10.1038/nsmb1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Esnault C, Ghavihelm Y, Brun S, Soutourina J, Vanberkum N, Boschiero C, Holstege F, Werner M. 2008. Mediator-dependent recruitment of TFIIH modules in preinitiation complex. Mol Cell 31:337–346. doi: 10.1016/j.molcel.2008.06.021. [DOI] [PubMed] [Google Scholar]
- 27.He Y, Yan C, Fang J, Inouye C, Tjian R, Ivanov I, Nogales E. 2016. Near-atomic resolution visualization of human transcription promoter opening. Nature 533:359–365. doi: 10.1038/nature17970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takagi Y, Komori H, Chang W-H, Hudmon A, Erdjument-Bromage H, Tempst P, Kornberg RD. 2003. Revised subunit structure of yeast transcription factor IIH (TFIIH) and reconciliation with human TFIIH. J Biol Chem 278:43897–43900. doi: 10.1074/jbc.C300417200. [DOI] [PubMed] [Google Scholar]
- 29.Murakami K, Gibbons BJ, Davis RE, Nagai S, Liu X, Robinson PJJ, Wu T, Kaplan CD, Kornberg RD. 2012. Tfb6, a previously unidentified subunit of the general transcription factor TFIIH, facilitates dissociation of Ssl2 helicase after transcription initiation. Proc Natl Acad Sci U S A 109:4816–4821. doi: 10.1073/pnas.1201448109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo J, Cimermancic P, Viswanath S, Ebmeier CC, Kim B, Dehecq M, Raman V, Greenberg CH, Pellarin R, Sali A, Taatjes DJ, Hahn S, Ranish J. 2015. Architecture of the human and yeast general transcription and DNA repair factor TFIIH. Mol Cell 59:794–806. doi: 10.1016/j.molcel.2015.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han Y, Luo J, Ranish J, Hahn S. 2014. Architecture of the Saccharomyces cerevisiae SAGA transcription coactivator complex. EMBO J 33:2534–2546. doi: 10.15252/embj.201488638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fan L, Arvai AS, Cooper PK, Iwai S, Hanaoka F, Tainer JA. 2006. Conserved XPB core structure and motifs for DNA unwinding: implications for pathway selection of transcription or excision repair. Mol Cell 22:27–37. doi: 10.1016/j.molcel.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 33.Sali A, Blundell TL. 1993. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 34.Lüthy R, Bowie JU, Eisenberg D. 1992. Assessment of protein models with three-dimensional profiles. Nature 356:83–85. doi: 10.1038/356083a0. [DOI] [PubMed] [Google Scholar]
- 35.Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen H-T, Warfield L, Hahn S. 2007. The positions of TFIIF and TFIIE in the RNA polymerase II transcription preinitiation complex. Nat Struct Mol Biol 14:696–703. doi: 10.1038/nsmb1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chin JW, Cropp TA, Anderson JC, Mukherji M, Zhang Z, Schultz PG. 2003. An expanded eukaryotic genetic code. Science 301:964–967. doi: 10.1126/science.1084772. [DOI] [PubMed] [Google Scholar]
- 38.Mohibullah N, Hahn S. 2008. Site-specific cross-linking of TBP in vivo and in vitro reveals a direct functional interaction with the SAGA subunit Spt3. Genes Dev 22:2994–3006. doi: 10.1101/gad.1724408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kellenberger E, Dominguez C, Fribourg S, Wasielewski E, Moras D, Poterszman A, Boelens R, Kieffer B. 2005. Solution structure of the C-terminal domain of TFIIH P44 subunit reveals a novel type of C4C4 ring domain involved in protein-protein interactions. J Biol Chem 280:20785–20792. doi: 10.1074/jbc.M412999200. [DOI] [PubMed] [Google Scholar]
- 40.Schmitt DR, Kuper J, Elias A, Kisker C. 2014. The Structure of the TFIIH p34 subunit reveals a von Willebrand factor A like fold. PLoS One 9:e102389. doi: 10.1371/journal.pone.0102389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kainov DE, Vitorino M, Cavarelli J, Poterszman A, Egly J-M. 2008. Structural basis for group A trichothiodystrophy. Nat Struct Mol Biol 15:980–984. doi: 10.1038/nsmb.1478. [DOI] [PubMed] [Google Scholar]
- 42.Iyer N, Reagan MS, Wu KJ, Canagarajah B, Friedberg EC. 1996. Interactions involving the human RNA polymerase II transcription/nucleotide excision repair complex TFIIH, the nucleotide excision repair protein XPG, and Cockayne syndrome group B (CSB) protein. Biochemistry 35:2157–2167. doi: 10.1021/bi9524124. [DOI] [PubMed] [Google Scholar]
- 43.Busso D, Keriel A, Sandrock B, Poterszman A, Gileadi O, Egly JM. 2000. Distinct regions of MAT1 regulate cdk7 kinase and TFIIH transcription activities. J Biol Chem 275:22815–22823. doi: 10.1074/jbc.M002578200. [DOI] [PubMed] [Google Scholar]
- 44.Rudolf J, Makrantoni V, Ingledew WJ, Stark MJR, White MF. 2006. The DNA repair helicases XPD and FancJ have essential iron-sulfur domains. Mol Cell 23:801–808. doi: 10.1016/j.molcel.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 45.Liu H, Rudolf J, Johnson KA, McMahon SA, Oke M, Carter L, McRobbie A-M, Brown SE, Naismith JH, White MF. 2008. Structure of the DNA repair helicase XPD. Cell 133:801–812. doi: 10.1016/j.cell.2008.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wolski SC, Kuper J, Hänzelmann P, Truglio JJ, Croteau DL, Van Houten B, Kisker C. 2008. Crystal structure of the FeS cluster-containing nucleotide excision repair helicase XPD. PLoS Biol 6:e149. doi: 10.1371/journal.pbio.0060149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fan L, Fuss JO, Cheng QJ, Arvai AS, Hammel M, Roberts VA, Cooper PK, Tainer JA. 2008. XPD helicase structures and activities: insights into the cancer and aging phenotypes from XPD mutations. Cell 133:789–800. doi: 10.1016/j.cell.2008.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]