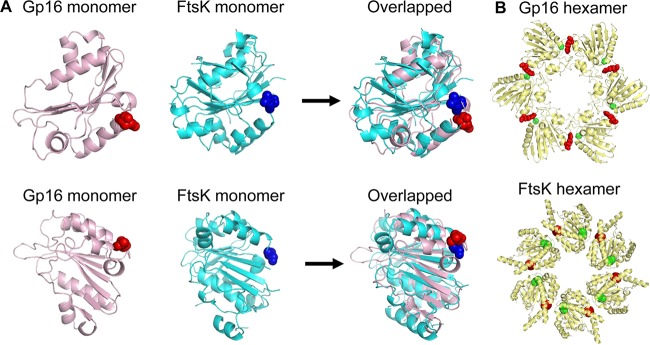
FIG 5.
Prediction and comparison of gp16 structure. (A) Structural comparison between the crystal structure of FtsK monomer (PDB accession number 2IUU; cyan) and the gp16 ATPase model (pink). The arginine finger is highlighted as a sphere. (B) Comparison of the predicted gp16 hexamer and FtsK hexamer. The ATPase gp16 hexamer structure was constructed using the predicted monomer structure shown in panel A and the P. aeruginosa FtsK (PDB accession number 2IUU) as templates (34). VMD was used to render the image of the structure (35). The ATP domains are highlighted as spheres: residue 27 (green, the conserved Walker ATP domain) and residue 146 (red, the arginine finger). The interaction of the arginine finger with the upstream adjacent subunit is evidenced by the proximity of the red and green spheres in both the constructed structure of the gp16 hexamer and the FtsK hexamer crystal structure.