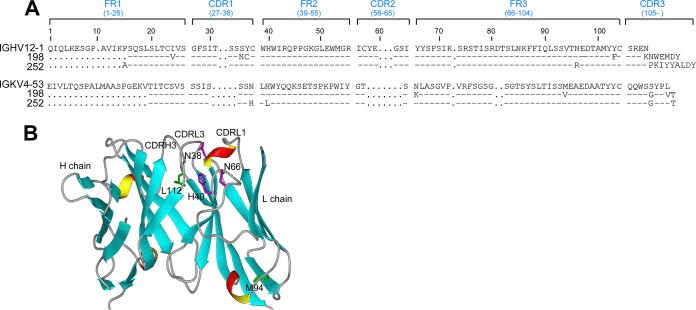
FIG 8.
Comparison of rMAbs 198 and 252. (A) Amino acid residue alignments of rMAbs 198 and 252. (B) Modeling of rMAb 198 with predicted IgLk residue differences from rMAb 252. Residues N/H38, H/L40, K/N66, V/M94, and V/L112 were modeled on an immunoglobulin variable region ribbon structure (heavy chain [PDB entry 1ACY] and light chain [PDB entry 5D8J], with homologous sequences). Residues V94 and V112, within FR3 and CDRL3, respectively, are illustrated as the homologous residues M94 and L112 in rMAb 252. Note that N38 and H40, located in CDRL1 and FR2, respectively, are adjacent to the CDRH3 and are replaced by H38 and L40 in rMAb 252. As illustrated, residue N66 at the beginning of FR3 is also near CDRL3 and is K66 in rMAb 198.