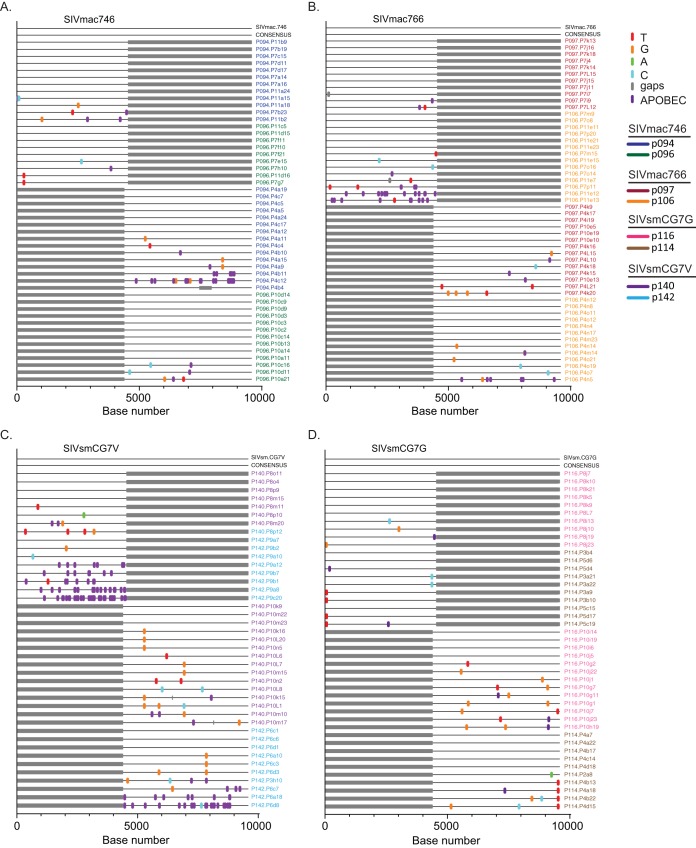
FIG 8.
Highlighter alignments of 5′ and 3′ half-genomes from all 8 infected animals. Plasma samples were obtained at peak viremia and used to identify the consensus genome for each infected animal. Each alignment represents two animals infected with each clone, SIVmac746 (A), SIVmac766 (B), SIVsmCG7V (C), and SIVsmCG7G (D). Single nucleotide polymorphisms to the T/F IMC sequences are denoted by colored tick marks; gaps are shown in gray and G-to-A mutations in purple. The full-genome consensus sequence (CONSENSUS) for both animals is an exact nucleotide match with that of the IMC clone used to infect each animal.