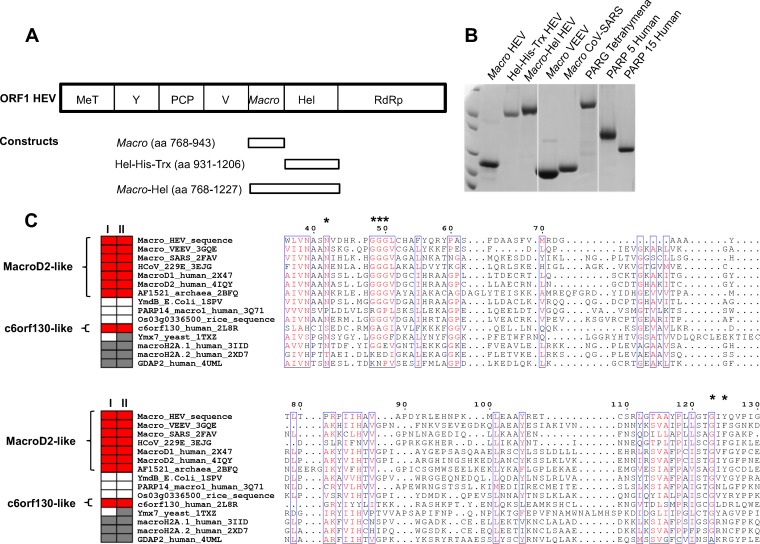
FIG 1.
Proteins relevant to this study. Predicted macro domain (amino acids [aa] 789 to 902) and Hel domain (amino acids 974 to 1185) inside ORF1 of the HEV genome (MeT, methyltransferase; Y, Y domain; PCP, papain-like cysteine protease; V, hypervariable proline-rich hinge; macro: macro domain; Hel, helicase; RdRP, RNA-dependent RNA polymerase). Rectangles show the produced proteins, as indicated. (B) Coomassie blue-stained SDS-PAGE gel showing the purified macro domain (macro-HEV), Hel (Hel-His-Trx HEV), macro-Hel, VEEV macro domain, SARS-CoV macro domain, Tetrahymena thermophila PARG, PARP5a catalytic domain (PARP5 human), and PARP15 catalytic domain (PARP15 human). (C) Structure-based sequence alignment of selected macro domains. Red rectangles, confirmed A1″Pase (I) or de-MARylation (II) activity; gray rectangles, no activity; blank rectangle, not tested. Activity data are based on the results of this study and previously published data (2). Macro domains carrying both A1″Pase and de-MARylation activities are further divided into MacroD2- and c6orf130-like subgroups, based on sequence conservation and catalytic mechanisms. Asterisks indicate important amino acids for PAR binding and/or de-MARylation activity of the MacroD2-like subgroup. E. coli, Escherichia coli; Trx, thioredoxin.