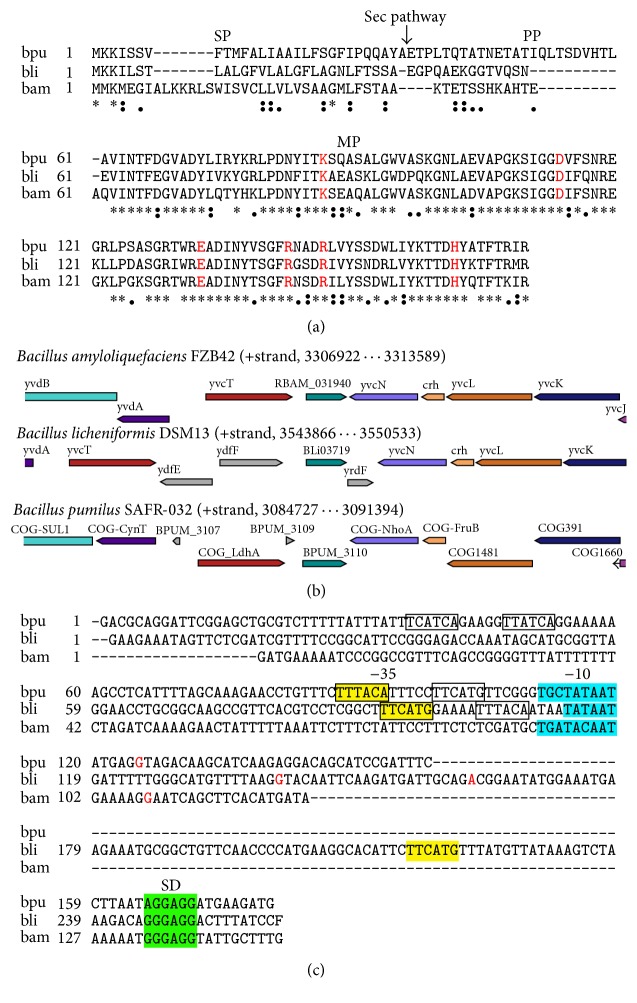
Figure 1.
Comparison of B. licheniformis RNase with the representatives of N1/T1 RNase family. (a) Sequence alignments of signal peptide (SP), propeptide (PP), and mature peptide (MP) of B. licheniformis (bli), B. amyloliquefaciens (bam), and B. pumilus (bpu) RNase. Identical amino acid residues are marked (∗). Amino acid residues that incorporate to the active site of enzyme are red colored. (b) Gene neighborhood of balifase gene in comparison to barnase and binase genes. The data were adopted from the MicrobesOnline Database (http://www.microbesonline.org/). (c) Promoter regions of guanyl-preferring RNase genes from B. pumilus (bpu), B. licheniformis (bli), and B. amyloliquefaciens (bam). Putative PhoP-binding sites are boxed; (+1) regions are red colored. A colon “:” indicates conservation between groups of strongly similar properties. A period “.” indicates conservation between groups of weakly similar properties.