Figure 4.
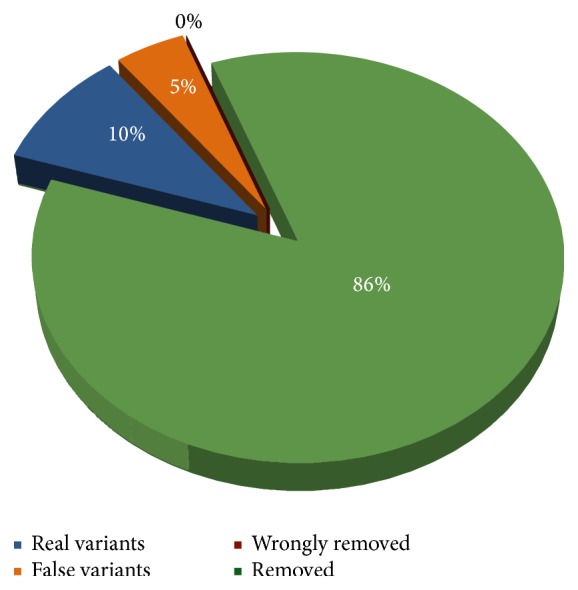
Performance of AGSA software for evaluation of homopolymers. 299 indel variants were found by AGSA in homopolymer sequences. After analysis of individual flowgrams, 246 (86%) were classified as false positive variants and 43 (14%) as true variants. Sanger sequencing confirmed that the 246 AGSA-classified false positives were actually wild-type sequences. Among the 43 potentially real variants, 29 (10%) were confirmed with Sanger analysis and 14 (5%) were actually wild-type Sanger sequences.
