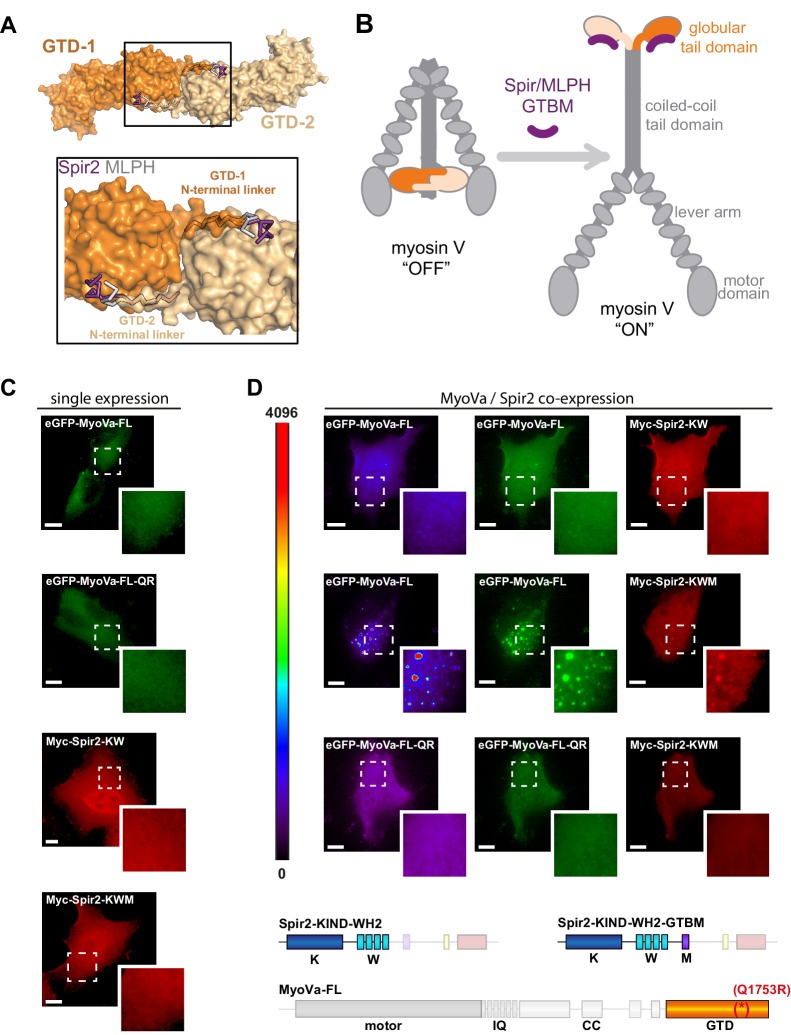
Figure 6. Spir-2 facilitates myosin V recruitment to vesicle membranes.
(A) A putative interaction mode of the GTDs within the MyoV off state was derived from the MyoVb-GTD structure (PDB ID 4LX0). Two GTDs interact with each other via N-terminal linkers occupying the Spir/MLPH binding pockets on the neighboring GTD, Spir and MLPH GTBMs are shown in ribbon. (B) Schematic representation of myosin V activation by GTBM binding. (C and D) N-terminal Spir fragments target MyoVa to vesicle membranes. (C) N-terminal Spir fragments (Myc-Spir-2-KIND-WH2, Myc-Spir-2-KW; Myc-Spir-2-KIND-WH2-GTBM, Myc-Spir-2-KWM) and full-length MyoVa motor proteins (eGFP-MyoVa-FL, eGFP-MyoVa-FL-Q1753R) have an even cytoplasmic and nuclear localization when transiently expressed in HeLa cells (single expression). At least 5 cells were recorded for each condition and one representative cell is presented here. Scale bars represent 10 µm. (D) Expression of full-length GFP-tagged MyoVa (eGFP-MyoVa-FL; green (middle) and as heat map (left)) shows an even cytoplasmic distribution that is not changed by co-expression of the Spir-2 fragment (Myc-Spir-2-KIND-WH2), which is not able to bind to MyoV and lipid membranes (upper panel). In contrast, co-expression of a myosin V binding Spir-2 fragment (Myc-Spir-2-KIND-WH2-GTBM; red, middle panel) leads to targeting of MyoVa to vesicle membranes and to overlapping Spir-2 and MyoVa localization (higher magnification insets). Heat maps represent grey values for MyoVa fluorescence intensities rising from '0' (black) to '4096' (red) to document equal expression levels of MyoVa proteins in the depicted cells. To address Rab11 dependence on motor protein targeting, the GFP-tagged melanocyte specific F isoform of the Q1753R mutant MyoVa (eGFP-MyoVa-QR) that does not bind Rab11 was expressed. The expressed MyoVa mutant has an even cytoplasmic distribution that was not changed upon co-expression of the myosin V binding Spir-2 fragment (Myc-Spir-2-KIND-WH2-GTBM; red, lower panel). Representative cells are shown. All cells observed under single and co-expression conditions had a vesicular or cytoplasmic localization as shown by the representative cells presented here. 5 cells were recorded for each condition and one representative cell is presented here. Scale bars represent 10 µm.