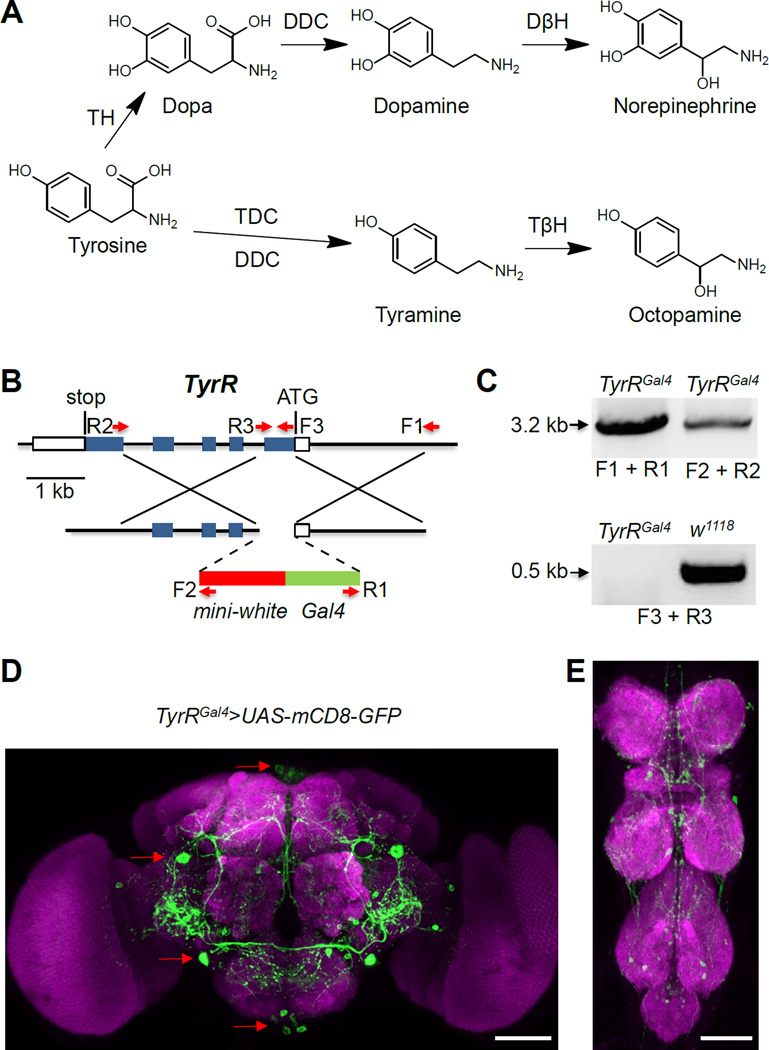
Figure 1. Synthesis of tyramine and knockout and expression of TyrR.
(A) Biosynthetic pathway of biogenic amines derived from tyrosine. DβH, dopamine β-hydroxylase; DDC, Dopa decarboxylase; TDC, tyrosine decarboxylase; TβH, tyramine β-hydroxylase; TH, tyrosine hydroxylase. Drosophila DDC is also capable of converting tyrosine to tyramine in vitro [49].
(B) Schematic of the TyrR locus (90C2-90C3) and the targeting construct used to generate the TyrRGal4 allele. The boxes represent exons and the coding regions are shown in blue. Indicated are the translational start codon (ATG), stop codon and the forward (F) and reverse (R) PCR primers.
(C) PCR confirmation of the deletion in TyrRGal4 and the replacement of the mini-white and Gal4 in TyrRGal4. We prepared genomic DNA and performed PCR using the primer pairs indicated in (B).
(D) UAS-mCD8-GFP expression driven under the control of the TyrRGal4/+ in the brain. The red arrows indicate the location of some of the neurons expressing the TyrR reporter. SMP, superior medial protocerebrum; PLP, posteriorlateral protocerebrum; IPS, inferior posterior slope; GNG, gnathal ganglia. MB, mushroom body; AL, antennal lobe; OL, optical lobe.
(E) TyrR-reporter expression in the ventral nerve cord. TG, thoracic ganglion; AG, abdominal ganglion.
The scale bars represent 50 µM.