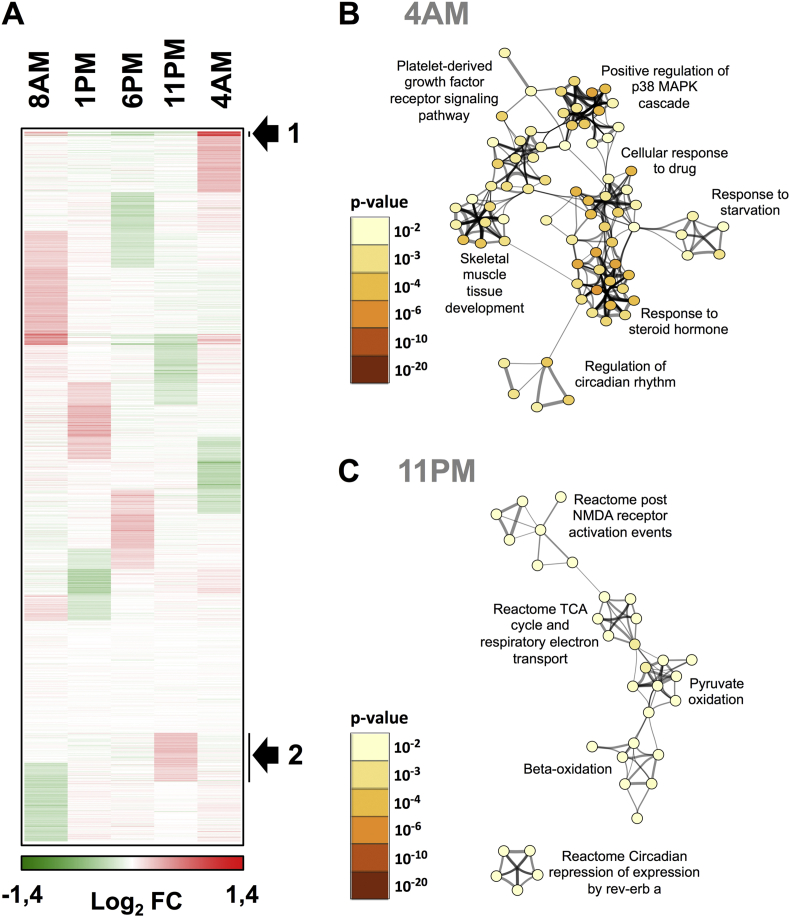
Figure 2.
Time-dependent expression of transcripts in human skeletal muscle. (A) Time-dependent gene expression was assessed by microarray analysis of RNA extracted from a single donor (N = 1). Protein-encoding transcript expression patterns were visualized as a self-organizing map (Euclidean distance metrics). The color scale indicates upregulated genes (red) and downregulated genes (green) relative to the global median signal of each array. The arrow indicates clusters of genes showing the most prominent peak of expression at the indicated time point (4 AM, cluster 1 and 11 PM, cluster 2). (B and C) Gene annotation enrichment analysis. The genes extracted from cluster 1 and 2 were searched using multiple databases (GeneOntology Biological Processes, KEGG pathways and Reactome) and statistically enriched terms were determined using the Metascape tool. The most significantly enriched terms are indicated for cluster 1 (GO biological processes, panel B, peak expression 4 AM) and cluster 2 (Reactome, panel C, peak expression 11 PM). Statistically significant terms were hierarchically clustered and converted into a network. Each term is represented by a circle node, of which the size is proportional to the number of genes in the term. The color of the node indicates the statistical significance of the term belonging to the cluster (see color scale). The most significant term characterizing each cluster is indicated. Similarities between terms are indicated by connecting lines.