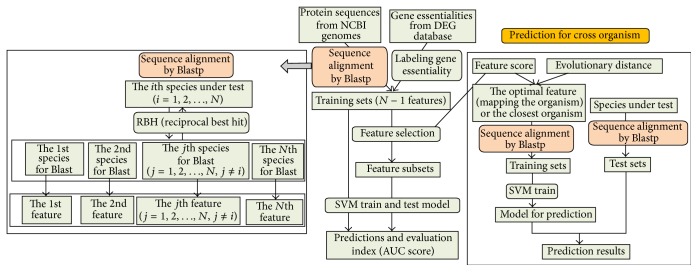
Figure 2.
The flowchart for obtaining training sets by multiple homology mapping and training the model to predict essential genes. For a species under test, it was used for sequence alignment towards other 24 species, respectively, and each result was used as a training feature. The training sets obtained from multiple sequence alignment were used to train and test the prediction model by SVM. Meanwhile, we used the F-score to evaluate the discriminative capability of each feature. The optimal feature subsets were selected to train and test the model. Tenfold cross-validation was utilized to assess the performance of the classifier. For predicting essential genes in cross organisms, the feature sets of the closest organism or those of the organism/feature which has the biggest F-score for the target species were selected as the training sets to train model, and then this model was used to predict essential genes in target species.