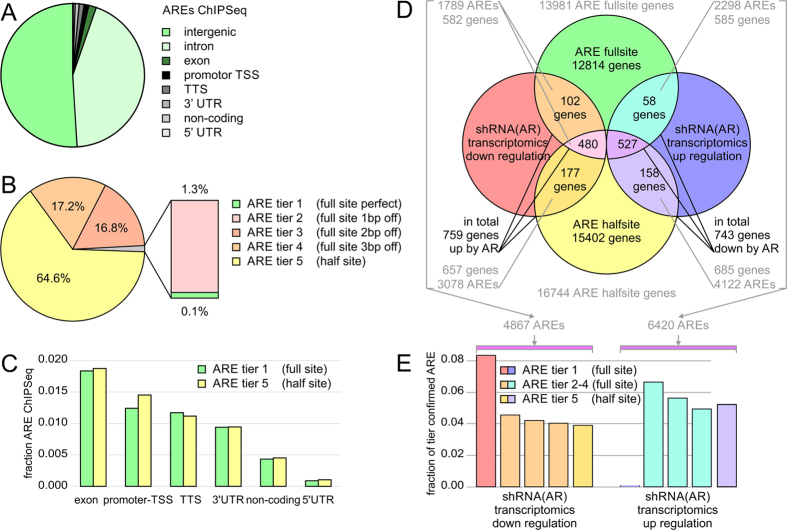
Figure 3. Genomic annotation and enrichment of AREs.
(A) Identified AREs in ChIP-Seq experiment were annotated by genomic elements. (B) Classification of AREs into five tiers. There were 71 AREs in tier 1 (perfect palindromic ARE), 1583 matches in tier 2 (1 bp off perfect), 20593 in tier 3 (2 bp off perfect), 21031 in tier 4 (3 bp off perfect) and 79065 in tier 5 (half site) with p-values below 4.94E-05 in the search for motif matches. (C) Genomic location of ARE full and half sites. Intergenic and intronic locations are not shown. (D) Overlay of gene mapping of AREs identified by ChIP-Seq and transcriptomics experiments. Using this data we defined the group of 759 genes as positively regulated by AR activity (down in cell with shRNA knockdown of AR), and 743 genes as negatively regulated by AR activity. (E) AREs mapped by ChIP-Seq experiments were confirmed by transcriptomics experiment and matched to genes. AREs confirmed by transcriptomics experiment showed higher hit rate in better defined tiers of AREs. Hit rate plotted as fraction of tier confirmed by significant down or up regulation in stable shRNA knockdown experiment with p-values below 0.05. Different tiers of AREs suggest different effect on transcriptional outcome as well as necessity of modulation of weaker motifs by coordinating factors. Imperfect AREs make up majority of genome-wide recognition motifs.