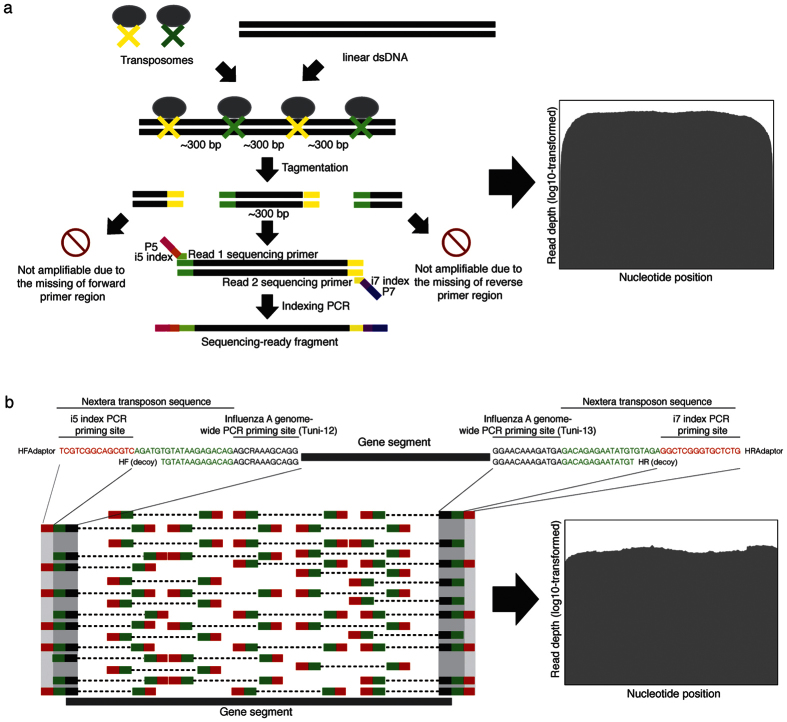
Figure 1. Optimization of the DNA library generation by introducing decoy PCR primers to produce full-length influenza A/H3N2 sequences with even coverage.
(a) The existing protocol for Nextera XT library preparation on targeted amplicons. Low sequencing read depths were typically found at both 5′ and 3′ ends of the amplicons. (b) The direct integration of the transposon sequences to the Tuni-12 and Tuni-13 primer sequences and inclusion of decoy primers during the initial genome-wide PCR before the Nextera XT library preparation. This increases the read depths at both ends of the amplicons.