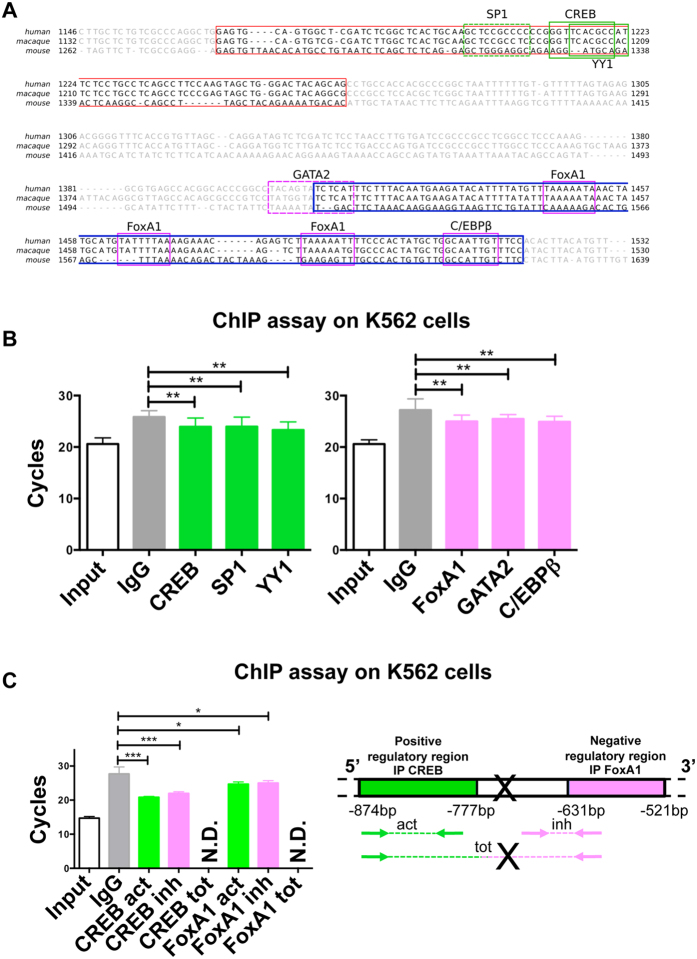
Figure 2. Identification of transcription factors and Chromatin Immuno Precipitation (ChIP) analysis.
(A) Conservation of identified activator and repressor sequence in human, macaque, and mouse. In red the region containing positive regulatory elements, from −874 to −777, in blue the region comprising negative regulatory elements, from −631 to −521. The location of putative binding sites for transcription factors are boxed: positive transcription factors are in green while negative are in pink. Solid line represent transcription factors bind to positive strand DNA, while dotted line indicate the ones bind to negative strand DNA. The sequence alignment was created with ClustalW2; transcription factors were identified by MatInspector and PROMO Alggen. (B) Chromatin immunoprecipitation (ChIP) prepared from K562 cells with the indicated set of antibodies specific for transcription factors selected by in silico analysis of the FTMT promoter. The amount of co-precipitated DNA is evaluated by qRT-PCR using oligonucleotides specific for positive and negative regulatory elements. On the left panel, ChIP of activator region: CREB, SP1 and YY1 transcription factors interacting with positive regulatory elements. On the right panel, ChIP of repressor region: FOXA1, GATA2 and C/EBPβ transcription factors interacting with negative regulatory elements. IgG represents the negative control with non-specific antibodies. Input represents the positive control of chromatin without immunoprecipitation. The plots represent the mean +/−SD of four independent experiments in triplicate, analyzed with One-way Anova with Bonferroni post test, **p < 0.01. (C) Chromatin immunoprecipitation (ChIP) as in B, the chromatin immunoprecipitated with the indicated transcription factors interacting with positive or negative regulatory elements was amplified also with the oligonucleotides specific for the opposite and the total regulatory elements (act = activator, inh = inhibitor, tot = total regulatory elements). N.D. = not detectable. X = possible broken region. The plots represent the mean +/−SD of two independent experiments in triplicate, analyzed with One-way Anova with Bonferroni post test, *p < 0.05, ***p < 0.001.