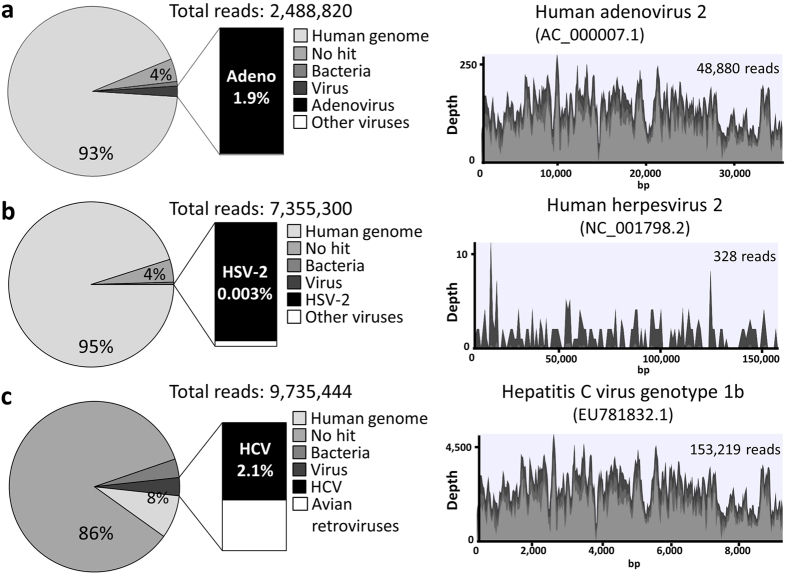
Figure 1. Identification of viral sequences in clinical samples using next-generation sequencing.
Pie charts show classification of reads from each library into human, unknown categories (no hit), bacteria and viruses. Total reads were mapped against the reference genomes using the CLC Genomics Workbench. Light gray, gray, and very light gray colors in the viral genome alignments represent minimal, average, and maximal coverage in the aggregated 100 bp (human adenovirus and hepatitis C virus) or 1 kbp (human herpesvirus 2) region, respectively. (a) Sequencing results of the DNA library prepared from the serum of a patient with adenovirus fulminant hepatitis. Sequencing reads were mapped to the reference genome of human adenovirus 2. (b) Sequencing results of the DNA library prepared from cerebrospinal fluid of a patient with neonatal herpes encephalitis. Sequencing reads were mapped to the reference genome of human herpesvirus 2. (c) Sequencing results of the RNA library prepared from the serum of a patient with hepatitis C virus infection. Sequencing reads were mapped to the reference genome of Hepatitis C virus genotype 1b.