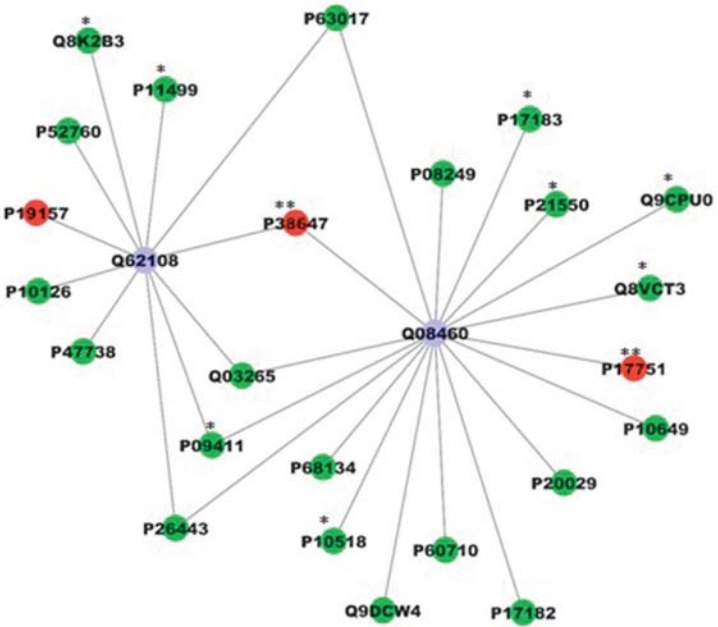
Figure 2. Subnetworks generated by VizMapper for each comparison – A Group A/J vs. 129p3/J. Color of node and * indicate the differential expression of the respective protein, for each comparison. Red and green nodes indicate protein down-regulation and up-regulation, respectively, while * and ** indicate presence and absence of protein, respectively, in the respective group. Purple node indicates proteins presenting interaction but that were not identified in the present study. The access numbers in nodes correspond to: P68134- (Acta1)Actin, alpha skeletal muscle; P10518- (Alad) Delta-aminolevulinic acid dehydratase; Q9DCW4- (Etfb) Electron transfer flavoprotein subunit beta; P60710- (Actb) Actin, cytoplasmic 1; P17182- (Eno1) Alpha-enolase; P20029- (Hspa5) 78 kDa glucose-regulated protein; P10649- (Gstm1) Glutathione S-transferase Mu 1; P17751- (Tpi1) Triosephosphate isomerase; Q8VCT3- (Rnpep) Aminopeptidase B; Q9CPU0- (Glo1) Lactoylglutathionelyase; P21550- (Eno3) Beta-enolase; P17183- (Eno2) Gamma-enolase; P08249- (Mdh2) Malate dehydrogenase; P63017- (Hspa8) Heat shock cognate; P38647- (Hspa9) Stress-70 protein; Q03265- (Atp5a1) ATP synthase subunit alpha; P09411- (Pgk1) Phosphoglycerate kinase 1; P26443- (Glud1) Glutamate dehydrogenase 1; P47738- (Aldh2) Aldehyde dehydrogenase; P10126- (Eef1a1) Elongation factor 1-alpha 1; P19157- (Gstp1) Glutathione S-transferase P 1; P52760- (Hrsp12) Ribonuclease; Q8K2B3- (Sdha) Succinate dehydrogenase; P11499- (Hsp90ab1) Heat shock protein; Q62108- (Dlg4) Disks large homolog 4; Q08460- (Kcnma1) Calcium-activated potassium channel subunit alpha-1.