ABSTRACT
Plant development depends on the perception of external cues, such as light, gravity, touch, wind or nutrients, among others. Nevertheless, little is known regarding signal transduction pathways integrating these stimuli. Recently, we have reported the involvement of a rice E3-ubiquitin ligase (OsHOS1, HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE1), previously associated with abiotic stress response, in root responses to mechanical stimuli. We showed that OsHOS1 is involved in the regulation of root curling after mechanosensing and that RNAi::OsHOS1 plants failed to exhibit the root curling phenotype observed in WT. Interestingly, the straight root phenotype of these transgenics correlated with the up-regulation of rice ROOT MEANDER CURLING (OsRMC, a negative regulator of rice root curling) and was reverted by the exogenous application of jasmonic acid. Altogether, our results highlight the role of the proteasome modulating plant responses to mechanical stimuli and suggest that OsHOS1 is a hub integrating environmental and hormonal signaling into plant growth and development.
KEYWORDS: E3-ubiquitin ligase, jasmonic acid, mechanosensing, rice, root
Due to their stationary nature and in order to optimize their growth, plants need to sense and continuously adjust their development through specific movements in response to the surrounding environment. These growth movements have called the attention of researchers for long (see Darwins's work in 1880). These movements are called tropisms and are named differently according to the nature of the stimulus (gravity – gravitropism; light – phototropism; touch – thigmotropism; etc.).1,2
The mechanical force-induced tropisms (e.g. wind, touch, gravity) are the most studied. The ability to perceive these forces seems to be essential to all plant cells, since even individual cells (e.g., protoplasts) have that capacity (see references herein1). Although it is very clear how plant tissues and cells respond to mechanical forces, the molecular mechanisms underlying these responses are still poorly understood. The large amount of genomic data and mutants made available over the last couple of decades unveiled some of the mechanisms involved. It seems clear that the signaling cascade triggered by the mechanical forces involves the modulation of ion fluxes (Ca2+, H+, osmolytes, ROS, etc.) and respective channels,3,4 most probably through receptor-like kinases.5 Perturbations in the structure of the cell-wall, plasma membrane and associated cytoskeleton, are also related to the initial stages of the tropic response to the mechanical forces.6 At transcriptional level, many genes have been shown to be regulated in response to mechanical forces,7,8 but several are also involved in other stress responsive pathways.8 These observations indicate that an extensive crosstalk is necessary to modulate mechanically-mediated growth in which plant hormones also play an important role.9,10
Besides transcriptional changes, the proteome remodelling by the ubiquitin/26S proteasome system (UPS) also plays an important role in the stress responses, allowing plants to increase their chances of survival.11 Over the last decade, the ubiquitin/26S proteasome system has emerged as an essential mechanism modulating plant hormonal signaling12 and also mediating abiotic and biotic stress responses.13 Ubiquitin is a small protein (8KDa) that is ubiquitous to all eukaryotic systems. Ubiquitin can be transferred to the target protein as a single molecule (mono-ubiquitination), multi-ubiquitin molecules or as an ubiquitin chain (polyubiquitination). The latter is the most common type of ubiquitination and it can prompt the ubiquitinated target protein for degradation through the 26S proteasome machinery.
High Expression of Osmotically Responsive Gene 1 (HOS1)
The E3-ubiquitin ligase HOS1 was initially identified in a mutant screen for cold responses in Arabidopsis. In the hos1 mutant, cold-responsive genes were up-regulated even without cold induction.14 Genes encoding the C-REPEAT/DEHYDRATION RESPONSIVE ELEMENT BINDING FACTOR (CBF/DREB1) transcription factors are up-regulated in the hos1 mutant, under control conditions, as compared to WT, thus accounting for the cold-responsive transcription pattern. The CBF/DREB1 gene expression is regulated by the HOS1 through the INDUCER OF CBF EXPRESSION 1 (ICE1) transcription factor, which directly regulates CBF/DREB1 gene expression. HOS1 interacts with ICE1, targeting this TF for degradation through the UPS, thus modeling cold responses.15
However, throughout the years, and despite the initial interest on HOS1 because of its involvement in cold stress signaling, it became evident to researchers that there were other interesting phenotypes of the hos1 mutant, thus revealing its putative involvement in other mechanisms. It was shown that HOS1 affects several plant processes, such as flowering time (through CONSTANS) and stomatal development (through ICE1/SCREAM). Other functions have been assigned to HOS1 besides its E3-ubiquitin ligase activity: as a member of the nucleopore complex, controlling import and export of mRNAs, controlling circadian rhythm components, or as a chromatin remodeller mediating FLOWERING LOCUS C expression. This places HOS1 as an important integrator of temperature and light signals, thus modulating developmental processes essential for plant survival (for reviews on Arabidopsis HOS1, see16,17 and references therein).
It is also important to state that these processes may also be controlled by plant hormones. Recently, it has been suggested that HOS1 is also involved in ethylene and auxins responses.18,19 Therefore, a cross-talk between the HOS1 associated processes and different plant hormones is expected.
Rice HOS1 (OsHOS1)
Previously, we have identified a rice RING E3-ubiquitin ligase, which was named rice HIGH OSMOTICALLY RESPONSIVE GENE 1 (OsHOS1) due to its sequence similarity with Arabidopsis HOS1 and ability to target OsICE1 for proteasomal degradation.20 In this work, we demonstrated that OsHOS1 interacts with OsICE1, thus modulating the expression of OsDREB1 genes. We observed an accumulation of OsICE1 protein in the rice RNAi::OsHOS1 line under control conditions, as compared to WT plants. Furthermore, using an in vitro degradation assay, we observed that tagged-OsICE1 protein was less degraded when incubated with total protein extracts from RNAi::OsHOS1 plants, as compared to WT.20 However, despite the accumulation of transcripts related to cold stress response and tolerance, the RNAi::OsHOS1 did not show enhanced cold tolerance as previously observed for the Arabidopsis hos1 mutant.14
As observed in Arabidopsis, the rice plants with lower HOS1 transcript levels showed pleiotropic phenotypes, besides the influence in the cold signaling pathway. In our recent work,21 we observed that RNAi::OsHOS1 plants have a straight root phenotype, as compared with WT (cv. Nipponbare) plants. This observation was striking as the WT plants, under the same growth conditions, showed a clear root curling.21 Throughout the years, some reports have shown the same root curling phenotype and hypothesized about the reasons of this behavior.22-24 Still, the question “why do rice roots curl?” remains unanswered. Recently, we have tested rice root vertical growth in order to hit a glass barrier, and clearly demonstrated that the root curling phenotype is a mechanosensing response to the physical barrier.21
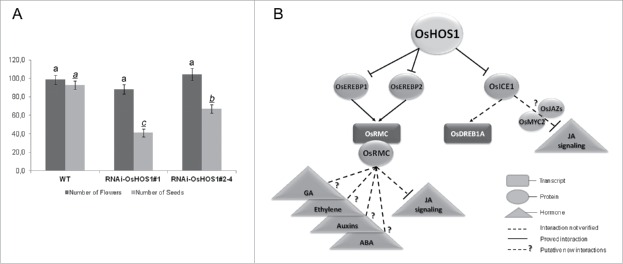
A correlation between the rice root curling phenotype and the Receptor-like kinase rice ROOT MEANDER CURLING (OsRMC) was previously reported,25 with OsRMC being described as a negative regulator of rice root curling through mechanisms involving jasmonic acid (JA). Therefore, we hypothesized that OsHOS1 could modulate OsRMC gene expression and impact rice root curling. In fact, when we tested OsRMC expression in WT and RNAi::OsHOS1 lines, we found that OsRMC expression was up-regulated in the transgenic plants and that we could establish a direct correlation between the frequency of the straight root phenotype and OsHOS1 transcript levels in the different transgenic lines. Furthermore, we showed that the root mechanosensing phenotype was correlated with JA. When we added JA to the medium, the roots of the transgenic RNAi::OsHOS1 lines were able to curl when challenged with a mechanical barrier. This confirmed the relation of OsRMC and JA as suggested by Jiang et al.25 At this point, we did not know if the effect over JA was at the level of its biosynthesis and/or of the signaling pathway. To assess this, we analyzed the expression of rice ALLENE OXIDE SYNTHASE 1 (OsAOS1), which encodes a key enzyme of the JA-biosynthesis pathway, and the classical effect of JA in root growth inhibition. Interestingly, the OsAOS1 expression remained unaltered in both lines analyzed (WT and RNAi::OsHOS1), while the root growth inhibition effect by JA was higher in WT roots than in RNAi::OsHOS1 plants. This result indicated a JA-signaling inhibition rather than changes in JA biosynthesis.21 Given that plants show a reduction in seed fertility when JA (biosynthesis or signaling) is affected26,27 we also assessed seed fertility in RNAi::OsHOS1 lines. When WT and transgenic plants were grown under semi-controlled conditions in a glasshouse (June-October 2015, Oeiras, Portugal), we found that, as compared to WT, RNAi::OsHOS1 lines showed identical number of flowers but a significant reduction in seed fertility (approximately 40% reduction), which correlated with the OsHOS1 transcript level (Fig. 1a). This result shows that OsHOS1 is involved in other developmental processes related to JA.
Figure 1.
Rice HOS1 as a putative regulator of developmental and stress-response pathways. (A) Graphical representation of the total number of flowers and total number of seeds per panicle of rice plants grown under semi-controlled conditions in a glasshouse (natural photoperiod between June-June–October 2015; Oeiras, Portugal). Glasshouse GPS coordinates N 38°41.770, W 009°19.247. Values represent means ± SE (n = 30 panicle per line). Statistical analysis was performed using ANOVA One-way analysis. The different letters above the bars (underlined letters for the number of seeds analysis) represent statistically significant differences between the different groups using Tukey's analysis (p < 0.01). (B) Schematic representation of the pivotal regulatory influence of OsHOS1. Lines represent proved interactions. Dashed lines represent influence not fully validated. Dashed lines with question mark (?) represent new hypothetical interactions.
Rice HOS1 REgulates two 2 ETHYLENE-RESPONSE FACTOR transcription factors
Previously, we had found two 2 ETHYLENE-RESPONSE FACTOR (ERF) transcription factors binding to the promoter of OsRMC.28 Therefore, we questioned whether the straight root phenotype could be related to a negative effect of OsHOS1 over the two 2 ERF (OsEREBP1 and OsEREBP2) transcription factors or directly to OsRMC. Being an E3-ubiquitin ligase, OsHOS1 could not directly affect OsRMC expression, but as seen above for Arabidopsis, other functions for this protein had to be taken into account. Our initial hypothesis was to test if OsHOS1 could interact directly with the two 2 ERF transcription factors binding to OsRMC promoter. Using Yeast Two-hybrid and Bimolecular Fluorescence Complementation, we proved the direct interaction of OsHOS1 with the two 2 ERF transcription factors.21 In addition, we showed that this interaction was related to the E3-ubiquitin ligase activity of OsHOS1. Using an in vitro degradation assay, we showed that OsEREBP1 and OsEREBP2 proteins were less degraded when incubated with RNAi::OsHOS1 total protein extracts than when incubated with WT protein extracts. In addition, when the extracts were incubated with a proteasomal inhibitor (MG132), the transcription factors degradation was delayed, thus showing the involvement of the UPS.
Discussion
Plants need to sense the surrounding environment to adjust their developmental processes in order to maximize growth. In our recent work, we showed how the E3-ubiquitin ligase OsHOS1 modulates rice root development in response to a mechanical barrier. We also showed that this response involves JA. In Arabidopsis, several plant hormones have been shown to influence root growth, development, and mechanosensing.10,29 Jasmonic acid has previously been shown to be involved in mechanosensing, mainly on the aerial part of plants.9 Only recently it was implicated in root growth and development in Arabidopsis.30 In rice, JA was shown to be involved in the regulation of auxin responsiveness in gravi-stimulated coleoptiles.31 These observations, together with our results, suggest that the regulation of root development by mechanical forces/stimuli may be conserved between monocots and dicots and involve JA-signaling.
Previously, other authors have hypothesized regarding the influence of light and touch in rice root mechanosensing response.23,24,32 Our work clearly showed that mechanosensing is able to induce rice root curling under light conditions, and provides insights on how this process is regulated at the molecular level. The obligatory presence of light to induce root curling may be related to JA. Jasmonic acid pathway is influenced by light33 and therefore may be necessary to induce root curling.
The relation of the ubiquitin/proteasome system with plant hormone signaling is known for several years.11,12 Most of the known plant hormones are regulated at the signaling level by the UPS. We found that the E3-ubiquitin ligase OsHOS1 is somehow involved in the regulation of JA-signaling, thus controlling root mechanosensing response. What is interesting in our findings is the fact that OsHOS1 does not seem to be regulating, directly, any of the known components of the JA-signaling pathway. It was previously reported that OsRMC is a negative regulator of JA-signaling.25 In fact, the OsHOS1 effect on JA-signaling might be correlated with its influence on OsRMC gene expression. Never-theless, and despite the evidences of the influence of OsRMC over JA-signaling, there is no evidence for its biological function. Few reports have shown the influence of OsRMC in developmental response but none of them reported its function.25,28,34,35 Thus, it is important to determine the biological function of this apoplastic protein as a negative regulator of JA-signaling or of any other signaling pathway (Fig. 1b). This is more important since, in the last years, some reports have shown a correlation/interaction between ICE1 protein and MYC2 and/or JASMONATE ZIM-DOMAIN (JAZ) repressors, which could interfere with the JA-signaling pathway.36,37 Therefore, and since RNAi::OsHOS1 plants have a higher amount of OsICE1,20 we cannot rule out the hypothesis that, besides OsRMC, OsICE1 also regulates JA-signaling. This hypothesis could place OsHOS1 as a pivotal regulator to balance the level of different plant hormones associated with stress and developmental responses, such as thigmomorphogenesis (Fig. 1b). Due to the extensive crosstalk between different plant hormones,38 this may be a serious possibility.
Conclusions
In our work,21 we have shown that the rice E3-ubiquitin ligase OsHOS1 is involved in the root mechanosensing by modulating OsRMC expression through the proteasomal degradation of two 2 ERF transcription factors. We cannot however rule out that other molecular mechanisms are also involved in root mechanosensing. The biological function of rice root curling is still poorly understood. We suggest that root curling could be a mechanosensing response allowing rice seedlings to establish in the soil, avoiding to be uprooted by heavy rains or flooding and thus an evolutionary advantage of this species.
Disclosure of potential confllicts of interest
No potential conflicts of interest were disclosed.
References
- 1.Monshausen GB, Haswell ES. A force of nature: molecular mechanisms of mechanoperception in plants. J Exp Bot 2013; 64:4663-80; PMID:23913953; http://dx.doi.org/ 10.1093/jxb/ert204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vandenbrink JP, Kiss JZ, Herranz R, Medina FJ. Light and gravity signals synergize in modulating plant development. Front Plant Sci 2014; 5; PMID:25389428; http://dx.doi.org/ 10.3389/fpls.2014.00563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hamilton ES, Schlegel AM, Haswell ES. United in diversity: mechanosensitive ion channels in plants. Ann Rev Plant Biol 2015; 66:113-37; PMID:25494462; http://dx.doi.org/25162526 10.1146/annurev-arplant-043014-114700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yuan F, Yang HM, Xue Y, Kong DD, Ye R, Li CJ, Zhang J, Theprungsirikul L, Shrift T, Krichilsky B. et al.. OSCA1 mediates osmotic-stress-evoked Ca2+ increases vital for osmosensing in Arabidopsis. Nature 2014; 514:367; PMID:25162526; http://dx.doi.org/ 10.1038/nature13593 [DOI] [PubMed] [Google Scholar]
- 5.Shih HW, Miller ND, Dai C, Spalding EP, Monshausen GB. The receptor-like kinase FERONIA is required for mechanical signal transduction in Arabidopsis seedlings. Curr Biol 2014; 24:1887-92; PMID:25127214; http://dx.doi.org/ 10.1016/j.cub.2014.06.064 [DOI] [PubMed] [Google Scholar]
- 6.Sampathkumar A, Yan A, Krupinski P, Meyerowitz EM. Physical forces regulate plant development and morphogenesis. Curr Biol 2014; 24:R475-R83; PMID:24845680; http://dx.doi.org/ 10.1016/j.cub.2014.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braam J. In touch: plant responses to mechanical stimuli. New Phytol 2005; 165:373-89; PMID:15720650; http://dx.doi.org/ 10.1111/j.1469-8137.2004.01263.x [DOI] [PubMed] [Google Scholar]
- 8.Figueiredo DD, Barros PM, Cordeiro AM, Serra TS, Lourenco T, Chander S, Oliveira MM, Saibo NJ. Seven zinc-finger transcription factors are novel regulators of the stress responsive gene OsDREB1B. J Exp Bot 2012; 63:3643-56; PMID:22412187; http://dx.doi.org/ 10.1093/jxb/ers035 [DOI] [PubMed] [Google Scholar]
- 9.Chehab EW, Yao C, Henderson Z, Kim S, Braam J. Arabidopsis touch-induced morphogenesis is jasmonate mediated and protects against pests. Curr Biol 2012; 22:701-6; PMID:22483939; http://dx.doi.org/ 10.1016/j.cub.2012.02.061 [DOI] [PubMed] [Google Scholar]
- 10.Lange MJP, Lange T. Touch-induced changes in Arabidopsis morphology dependent on gibberellin breakdown. Nature Plants 2015; 1:14025; PMID:27246879; http://dx.doi.org/ 10.1038/nplants.2014.25 [DOI] [PubMed] [Google Scholar]
- 11.Dreher K, Callis J. Ubiquitin, hormones and biotic stress in plants. Ann Bot 2007; 99:787-822; PMID:17220175; http://dx.doi.org/ 10.1093/aob/mcl255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kelley DR, Estelle M. Ubiquitin-mediated control of plant hormone signaling. Plant Physiol 2012; 160:47-55; PMID:22723083; http://dx.doi.org/ 10.1104/pp.112.200527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dametto A, Buffon G, Dos Reis Blasi EA, Sperotto RA. Ubiquitination pathway as a target to develop abiotic stress tolerance in rice. Plant Signal Behav 2015; 10:e1057369; PMID:26236935; http://dx.doi.org/ 10.1080/15592324.2015.1057369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee HJ, Xiong LM, Gong ZZ, Ishitani M, Stevenson B, Zhu JK. The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold-regulated nucleo-cytoplasmic partitioning. Genes Dev 2001; 15:912-24; PMID:11297514; http://dx.doi.org/ 10.1101/gad.866801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong CH, Agarwal M, Zhang YY, Xie Q, Zhu JK. The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc Natl Acad Sci USA 2006; 103:8281-6; PMID:16702557; http://dx.doi.org/ 10.1073/pnas.0602874103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jung JH, Lee HJ, Park MJ, Park CM. Beyond ubiquitination: proteolytic and nonproteolytic roles of HOS1. Trends Plant Sci 2014; 19:538-45; PMID:24768209; http://dx.doi.org/ 10.1016/j.tplants.2014.03.012 [DOI] [PubMed] [Google Scholar]
- 17.MacGregor DR, Penfield S. Exploring the pleiotropy of hos1. J Exp Bot 2015; 66:1661-71; PMID:25697795; http://dx.doi.org/ 10.1093/jxb/erv022 [DOI] [PubMed] [Google Scholar]
- 18.Lee K, Seo PJ. The Arabidopsis E3 ubiquitin ligase HOS1 contributes to auxin biosynthesis in the control of hypocotyl elongation. Plant Growth Regul 2015; 76:157-65; http://dx.doi.org/ 10.1007/s10725-014-9985-x [DOI] [Google Scholar]
- 19.Lee K, Seo PJ. The E3 ubiquitin ligase HOS1 is involved in ethylene regulation of leaf expansion in Arabidopsis. Plant Signal Behav 2015; 10:e1003755; PMID:25848954; http://dx.doi.org/ 10.1080/15592324.2014.1003755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lourenço T, Sapeta H, Figueiredo DD, Rodrigues M, Cordeiro A, Abreu IA, Saibo NJ, Oliveira MM. Isolation and characterization of rice (Oryza sativa L.) E3-ubiquitin ligase OsHOS1 gene in the modulation of cold stress response. Plant Mol Biol 2013; 83:351-63; PMID:23780733; http://dx.doi.org/ 10.1007/s11103-013-0092-6 [DOI] [PubMed] [Google Scholar]
- 21.Lourenço TF, Serra TS, Cordeiro AM, Swanson SJ, Gilroy S, Saibo NJM, Oliveira MM. The rice E3-ubiquitin ligase HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE1 modulates the expression of ROOT MEANDER CURLING, a gene involved in root mechanosensing, through the interaction with two ETHYLENE-RESPONSE FACTOR transcription factors. Plant Physiol 2015; 169:2275-87; PMID:26381316; http://dx.doi.org/ 10.1104/pp.15.01131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimizu H, Tanabata T, Xie XZ, Inagaki N, Takano M, Shinomura T, Yamamoto KT. Phytochrome-mediated growth inhibition of seminal roots in rice seedlings. Physiol Plant 2009; 137:289-97; PMID:197-44160; http://dx.doi.org/ 10.1111/j.1399-3054.2009.01277.x [DOI] [PubMed] [Google Scholar]
- 23.Wang SJ, Ho CH, Chen HW. Rice develop wavy seminal roots in response to light stimulus. Plant Cell Reports 2011; 30:1747-58; PMID:21573806; http://dx.doi.org/ 10.1007/s00299-011-1082-2 [DOI] [PubMed] [Google Scholar]
- 24.Wang SJ, Kang CH, Chen HW. Effect of the interaction between light and touch stimuli on inducing curling seminal roots in rice seedlings. Plant Signal Behav 2011; 6:1434-5; PMID:21912213; http://dx.doi.org/ 10.4161/psb.6.10.17087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang JF, Li JH, Xu YY, Han Y, Bai Y, Zhou GX, Lou Y, Xu Z, Chong K. RNAi knockdown of Oryza sativa root meander curling gene led to altered root development and coiling which were mediated by jasmonic acid signalling in rice. Plant Cell Environm 2007; 30:690-9; PMID:17470145; http://dx.doi.org/ 10.1111/j.1365-3040.2007.01663.x [DOI] [PubMed] [Google Scholar]
- 26.Bae HK, Kang HG, Kim GJ, Eu HJ, Oh SA, Song JT, Chung IK, Eun MY, Park SK. Transgenic rice plants carrying RNA interference constructs of AOS (allene oxide synthase) genes show severe male sterility. Plant Breed 2010; 129: 647–51; http://dx.doi.org/ 10.1111/j.1439-0523.2010.01784.x [DOI] [Google Scholar]
- 27.Xiao YG, Chen Y, Charnikhova T, Mulder PPJ, Heijmans J, Hoogenboom A, Agalou A, Michel C, Morel JB, Dreni L. OsJAR1 is required for JA-regulated floret opening and anther dehiscence in rice. Plant Mol Biol 2014; 86:19-33; PMID:24947835; http://dx.doi.org/ 10.1007/s11103-014-0212-y [DOI] [PubMed] [Google Scholar]
- 28.Serra TS, Figueiredo DD, Cordeiro AM, Almeida DM, Lourenço T, Abreu IA, Sebastián A, Fernandes L, Contreras-Moreira B, Oliveira MM, et al.. OsRMC, a negative regulator of salt stress response in rice, is regulated by two AP2/ERF transcription factors. Plant Mol Biol 2013; 82:439-55; PMID:23703395; http://dx.doi.org/ 10.1007/s11103-013-0073-9 [DOI] [PubMed] [Google Scholar]
- 29.Massa GD, Gilroy S. Touch modulates gravity sensing to regulate the growth of primary roots of Arabidopsis thaliana. Plant J 2003; 33:435-45; PMID:12581302; http://dx.doi.org/ 10.1046/j.13-65-313X.2003.01637.x [DOI] [PubMed] [Google Scholar]
- 30.Gasperini D, Chetelat A, Acosta IF, Goossens J, Pauwels L, Goossens A, Dreos R, Alfonso E, Farmer EE. Multilayered organization of jasmonate signalling in the regulation of root growth. PloS Genet 2015; 11; PMID:26070206; http://dx.doi.org/ 10.1371/journal.pgen.1005300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gutjahr C, Riemann M, Muller A, Duchting P, Weiler EW, Nick P. Cholodny-Went revisited: a role for jasmonate in gravitropism of rice coleoptiles. Planta 2005; 222:575-85; PMID:16047199; http://dx.doi.org/ 10.1007/s00425-005-0001-6 [DOI] [PubMed] [Google Scholar]
- 32.Chen HW, Shao KH, Wang SJ. Light-mediated modulation of helix angle and rate of seminal root tip movement determines root morphology of young rice seedlings. Plant Signal Behav 2016:0 PMID:26829414; http://dx.doi.org/24014863 10.1080/15592324.2016.1141861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Svyatyna K, Riemann M. Light-dependent regulation of the jasmonate pathway. Protoplasma 2012; 249:137-45; PMID:22569926; http://dx.doi.org/24014863 10.1007/s00709-012-0409-3 [DOI] [PubMed] [Google Scholar]
- 34.Yang A, Li Y, Xu Y, Zhang WH. A receptor-like protein RMC is involved in regulation of iron acquisition in rice. J Exp Bot 2013; 64:5009-20; PMID:24014863; http://dx.doi.org/ 10.1093/jxb/ert290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang L, Tian LH, Zhao JF, Song Y, Zhang CJ, Guo Y. Identification of an apoplastic protein involved in the initial phase of salt stress response in rice root by two-dimensional electrophoresis. Plant Physiol 2009; 149:916-28; PMID:19036832; http://dx.doi.org/ 10.1104/pp.108.131144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu YR, Jiang LQ, Wang F, Yu DQ. Jasmonate regulates the INDUCER OF CBF EXPRESSION-C-REPEAT BINDING FACTOR/DRE BINDING FACTOR1 cascade and freezing tolerance in Arabidopsis. Plant Cell 2013; 25:2907-24; PMID:23933884; http://dx.doi.org/ 10.1105/tpc.113.112631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao ML, Wang JN, Shan W, Fan JG, Kuang JF, Wu KQ, Li XP, Chen WX, He FY, Chen JY, et al.. Induction of jasmonate signalling regulators MaMYC2s and their physical interactions with MaICE1 in methyl jasmonate-induced chilling tolerance in banana fruit. Plant Cell Environm 2013; 36:30-51; PMID:22651394; http://dx.doi.org/ 10.1111/j.1365-3040.2012.02551.x [DOI] [PubMed] [Google Scholar]
- 38.Song SS, Qi TC, Wasternack C, Xie DX. Jasmonate signaling and crosstalk with gibberellin and ethylene. Curr Opin Plant Biol 2014; 21:112-9; PMID:25064075; http://dx.doi.org/ 10.1016/j.pbi.2014.07.005 [DOI] [PubMed] [Google Scholar]