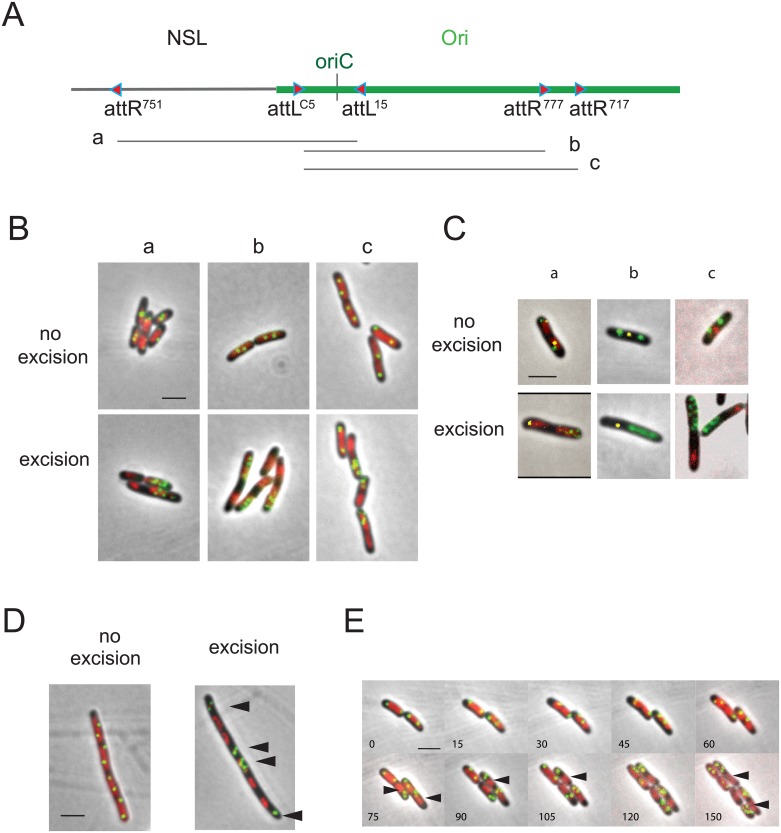
Fig 6. E. coli nucleoid positioning and chromosome segregation.
(A) The extent of the excised segment is indicated on the diagram below the map of the NSL-Ori region (segments a, b and c). (B) Montage of merged pictures of parSP1 (green), DAPI staining (red) and phase-contrast micrographs (grey) of MG1655 cells upon excision of chromosomal DNA segments a, b and c carrying oriC and a parS (Ori-5 or Ori-6). Control sample in the absence of excision is presented on the top panels. (C) Montage of merged pictures of MG1655 cells upon excision of chromosomal DNA segments carrying oriC and the parS tag (Ori-5 and Ori-6; indicated in green), together with a parST1 (indicated in yellow) on the remnant part either in Ori region (panel a) or in Ter region (panel b) or expressing a MatP-mCherry fusion protein (panel c), HU-mCherry staining (red; panel a) and phase-contrast micrographs (grey). Control sample in the absence of excision is presented on the top panels. (D) Montage of merged pictures of parSP1 Ori-6 (green), DAPI staining (red) and phase-contrast micrographs (grey) of MG1655 cells upon excision of chromosomal DNA segment b carrying oriC and the parS tag (Ori-6) in the presence of cephalexin. Control sample in the absence of excision is presented. Black arrowheads indicate excised rings carrying Ori markers. (E) Time-lapse experiment representing the dynamics of parSP1 Ori-6 tag upon excision of segment b. Montage of merged pictures of MG1655 cells upon excision of chromosomal DNA segments carrying oriC and the parS tag (Ori-6) and expressing HU-mCherry. Positioning of the focus was observed for 300 min with 10 min intervals. Black arrowheads indicate chromosome remnants. Scale bar indicate 2 μm.