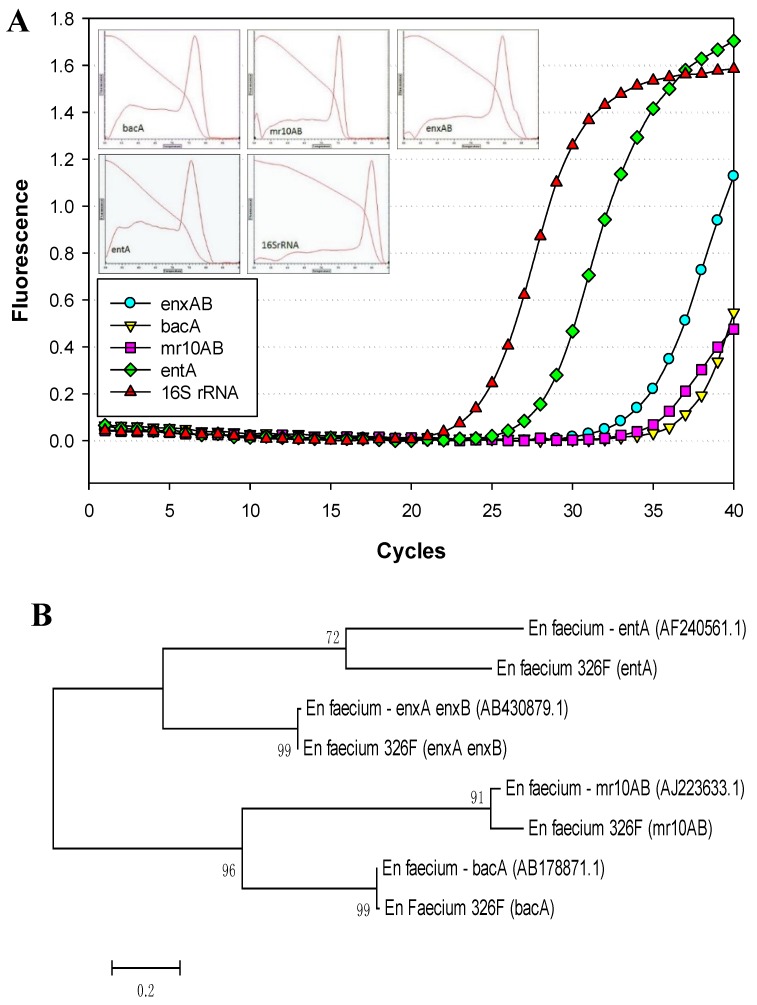
Figure 2.
Real-time PCR and dendrogram of sequence alignment of four bacteriocins from En. faecium 326F. Panel A, real-time PCR amplification of En. faecium 326F using a PCR array of 16 primer pairs (in individual reactions) for known enterococcal bacteriocins, including a 16S rRNA control. Only positive amplification reactions are shown and include primers derived from enterocins enxAB, mr10AB, entA, bacA, and universal 16S rRNA; the 12 other primer sets did not amplify. Insets are for melting curve assays of the positive amplicons (as indicated). Panel B, maximum likelihood homology tree obtained from multiple sequence alignment and phylogenetic analysis of four bacteriocin-related sequences obtained with En. faecium 326F (entA, mr10AB, enxAB, bacA) vs. the GenBank bacteriocin sequences from which the primers were derived: AF240561.1 (entA), AJ223633.1 (mr10AB), AB430879.1 (enxAB), and AB178871.1 (bacA).