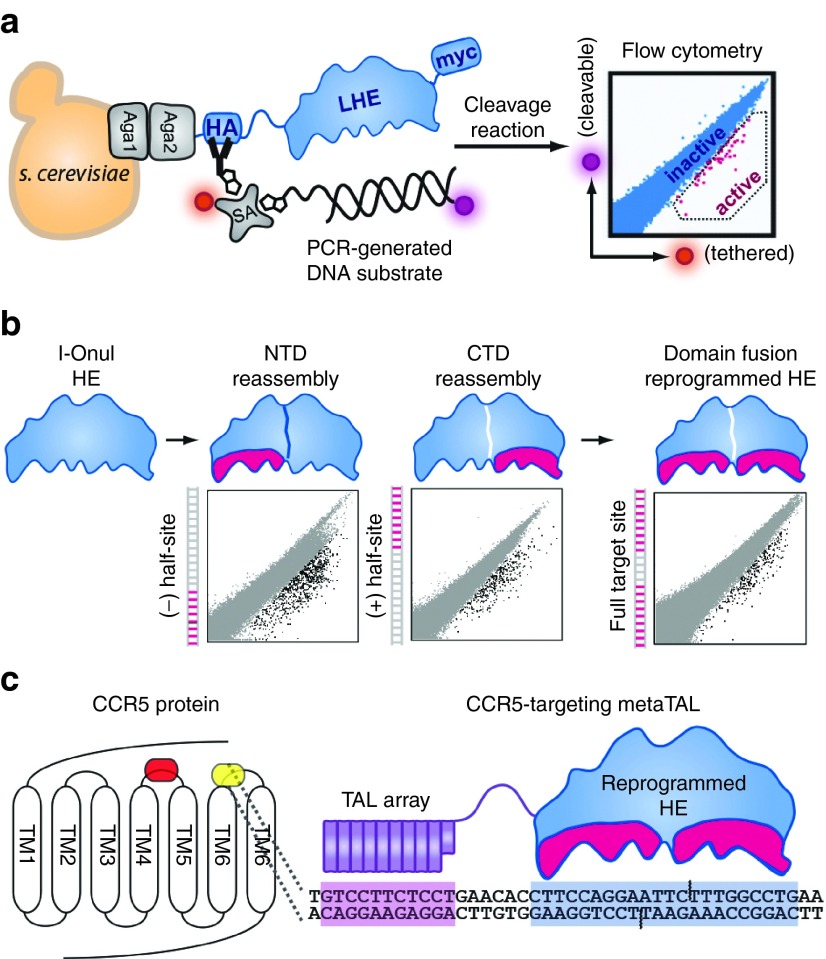
Figure 1.
Assembly of a reprogrammed megaTAL targeting the CCR5 gene. (a) Schematic depiction of yeast surfaced display technology used to reprogram the specificity of LAGLIDADG-motif homing endonuclease (LHE) I-OnuI. (b) The N-terminal (NTD) and C-terminal domains (CTD) of the LHE component of the megaTAL were reprogrammed separately to achieve activity against 9-base pair “half-sites” that were subsequently assembled to produce the fully reprogrammed enzyme recognizing the complete CCR5 target site. (c) CCR5 is a G-protein coupled receptor with seven transmembrane (TM) domains whose extracellular facing loops comprise, in part, the binding interface used by HIV-1 to gain entry to CD4+ T cells. We targeted the third extracellular loop (ECL3) located between the sixth (TM6) and seventh (TM7) transmembrane domains (yellow shading), downstream of the CCR5Δ32 deletion (orange shading). This region was selected for disruption, as this loop contains a cysteine residue that contributes to the structural integrity of CCR5 and its ability to function as an HIV-1 coreceptor.52 The megaTAL targeting the CCR5 ECL3 loop is shown schematically, with a modular transcription activator-like effector array (purple) linked to the reprogrammed I-OnuI LHE (HE; blue) and their corresponding target sequences separated by 6 base pairs of noncontacted DNA. The amino acid sequence for the CCR5 megaTAL has been previously published.25 Aga1 and Aga2, subunits of a-agglutinin heterodimer; HA, hemagglutinin; SA, streptavidin; Myc, c-myc tag.