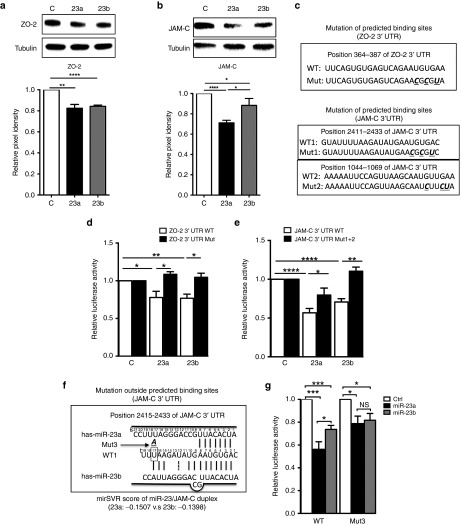
Figure 5.
MiR-23 directly target ZO-2 and JAM-C. (a) overexpression of miR-23a and miR-23b inhibited ZO-2 protein expression to a similar extent, n = 4 independent experiments.(b) overexpression of miR-23a inhibited JAM-C to a greater extent than miR-23b overexpression, n = 7 independent experiments. Representative Western blot is shown in upper panel and quantified by ImageJ as Pixel density. α-Tubulin was used as a loading control. (c) Putative miR-23 binding site and mutation within the ZO-2 and JAM-C 3′UTR. Nucleotides that were mutated are given in bold, underlined and italic ones in Mut. (d,e) Luciferase reporter assays, in which HEK293T cells were cotransfected with reporter constructs containing full length 3′UTR of the wild-type ZO-2, JAM-C mRNA (WT) or mutated miR-23 binding sites (Mut), together with miR-23a, miR-23b or control mimics. n = 3 independent experiments. (f) Schematic alignment between mature miR-23a/b and JAM-C 3′UTR shown at www.microrna.org. g, guide miRNA; t, target mRNA. Site t17 (in the rectangle) within WT1 was mutated to A (bold, italic and underlined) to become Mut3. (g) Luciferase reporter assays, in which HEK293T cells were cotransfected with reporter constructs containing full length 3′UTR of the wild-type JAM-C (WT), or single nucleotide mutation outside binding sites (Mut3), together with miR-23a, miR-23b or control mimics. n = 7 independent experiments. Data in all graph shown as mean ± SEM. *P < 0.05; **P < 0.01; ****P < 0.0001.