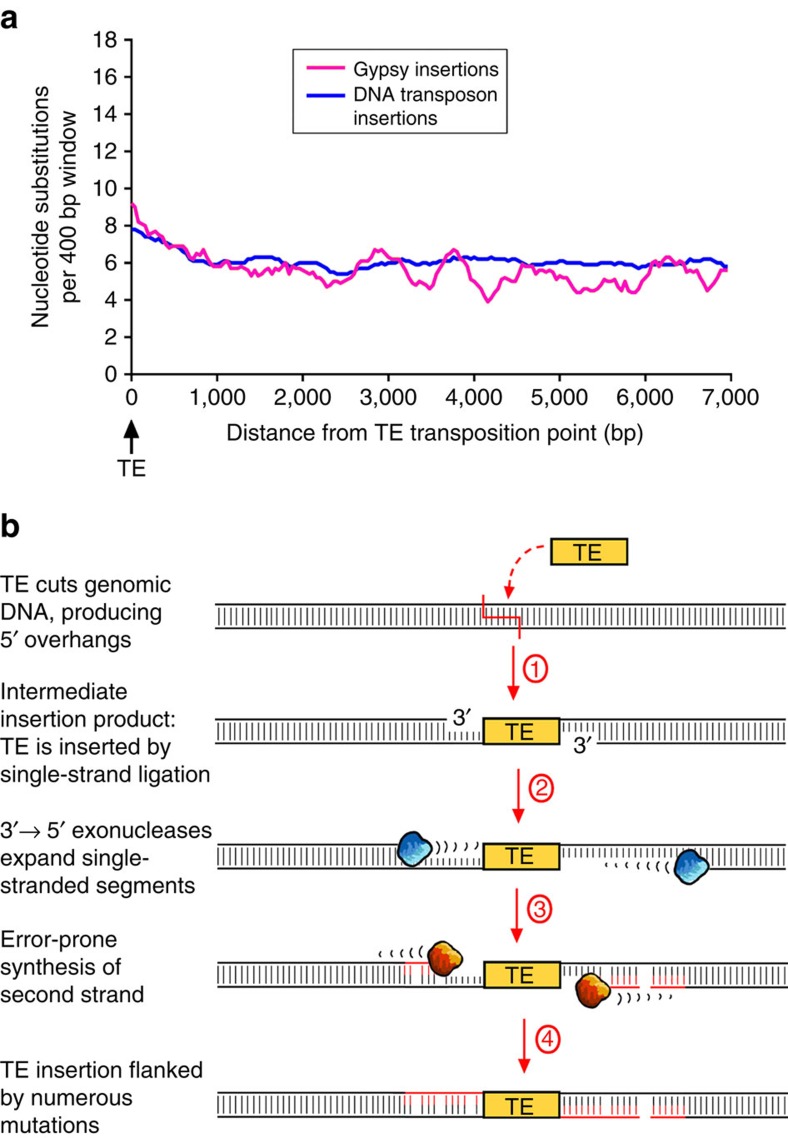
Figure 3. Mutations in sequences flanking transposable element insertions.
(a) Frequencies of nucleotide substitutions relative to insertion sites of DNA transposons and Gypsy retrotransposons sites in rice. In both cases, nucleotide substitution frequency increases slightly towards the insertion point. This indicates that insertions are also associated with small numbers of mutations in their flanking sequences. Furthermore, this result is evidence that events classified as DNA transposon insertions probably do not contain many precise excisions. (b) Proposed mechanism for error-prone DNA repair following TE insertions (see also Supplementary Fig. 3). Step 1: the TE inserts into the genome by producing a staggered cut, resulting in a TE that is ligated to the genomic DNA via single-stranded segments. Step 2: 3′→5′ exonucleases expose large stretches of single-stranded DNA. Step 3: second strands are synthesized by a replication complex that has deficiencies in DNA polymerases fidelity and mismatch repair (the same as described in Fig. 2c). Step 4: the final TE insertion is flanked by segments rich in nucleotide substitutions and small insertions and deletions.