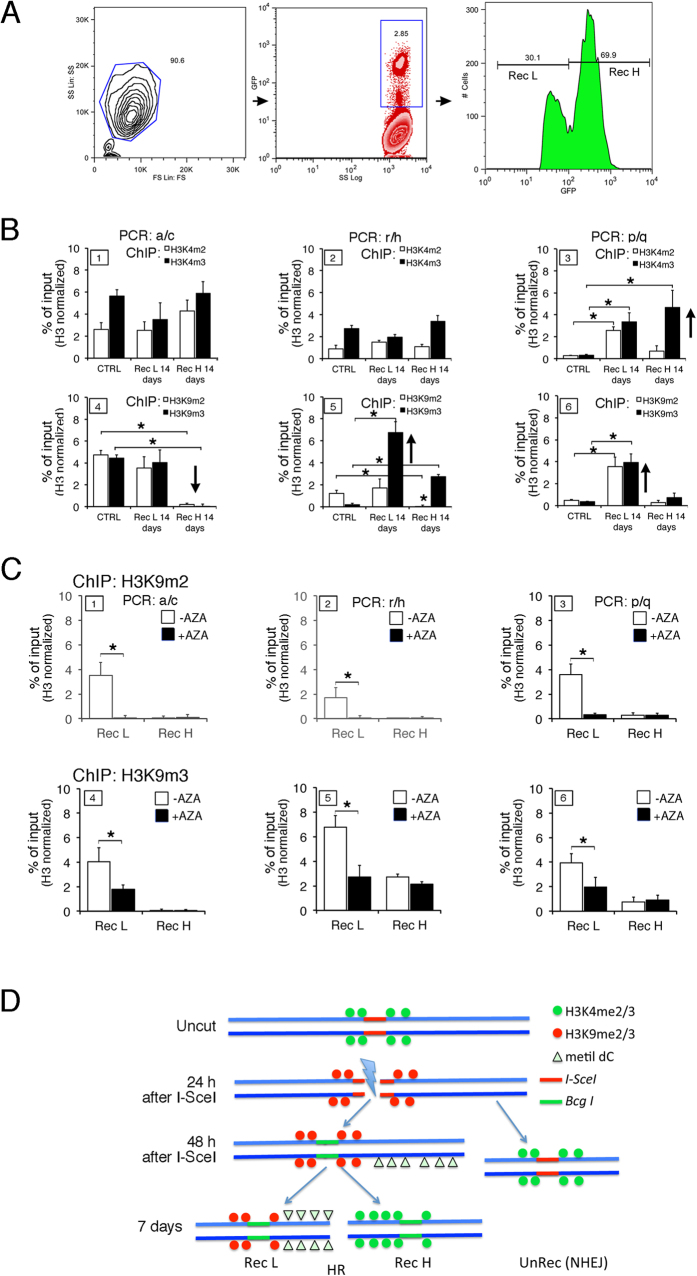
Figure 2. Spatial and temporal changes of histone H3 K4-K9 methylation after homologous repair at the GFP locus in HR cells.
(A) DRGFP Hela cells were transfected with the I-SceI vector and sorted for GFP expression 14 days later as described in Methods. The panels from the left to the right show the gating strategy used to sort Rec H and Rec L cells. The percent of viable cells was 90.6. The fraction of the total GFP+ cells (middle panel) or of Rec H and Rec L (left panel) is indicated. (B) H3K4m2/3 and H3K9m2/3 levels at GFP chromatin in purified Rec H and Rec L cells, 14 days after I-SceI transfection. The specific primers are indicated at the top of each column as shown in Fig. 1A. The data are normalized to total H3 content. *P < 0.01 (t test) as compared with cells transfected with control plasmid (CTRL). (C) H3K9m2/3 content of GFP in chromatin derived from sorted Rec H or Rec L clones, untreated or treated with 5-azadC (10 μM for 3 days and analyzed 4 days later). ChIP analysis was performed with the indicated antibodies. qPCR on each immunoprecipitate was carried out with primers r/h, as shown in Fig. 1A. *p < 0.01 (Wilcoxon rank-sum test) as compared with untreated control or (primers a/c). (D) Histone H3 and DNA methylation changes following the DSB and HR, A schematic cartoon illustrating the major chromatin changes of H3K4me2/3 or H3K9me2/3 and DNA methylation (Δ) following the DSB and the HR or NHEJ in recombinant (HR) and non recombinant (NHEJ) cells. The detailed statistical analysis of the data shown in panels (B,C) is reported in Supplemental Statistical Tables 3 and 4.