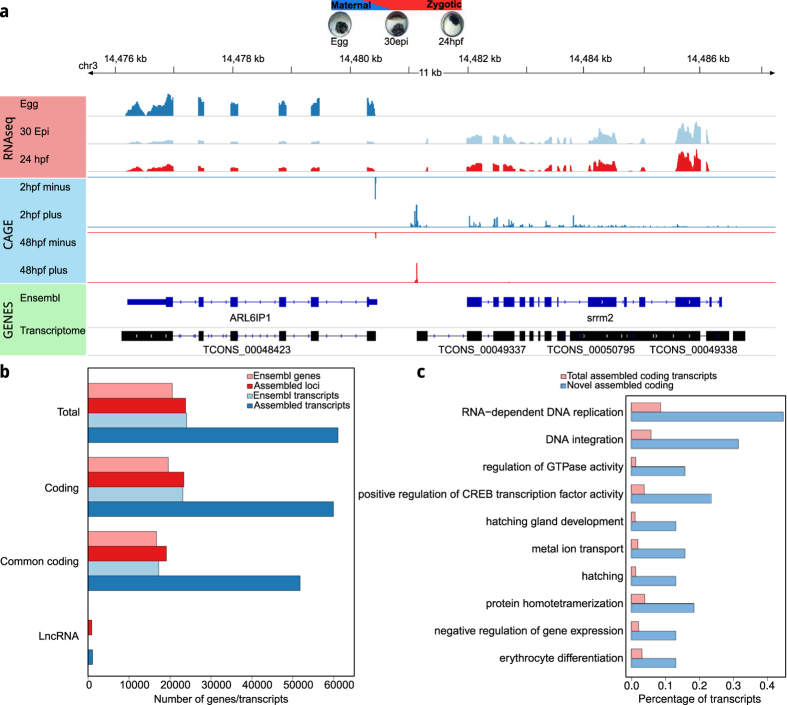
Figure 1. Principal features of the assembled developmental transcriptome of Tetraodon nigroviridis.
(a) Coverage plot from a representative 12 KB region on the tetraodon genome showing data from RNAseq of the three developmental stages, CAGE transcriptional start site peaks, Ensembl genes and assembled transcript models. (b) Comparison of the number of assembled loci and transcripts against the existing gene models in Ensembl. Common coding refers to assembled loci and transcripts mapped to a known Ensembl transcript model. (c) GO enrichment of the novel discovered coding transcripts. The novel coding transcripts are enriched in GO terms deemed important for early embryo development. The x-axis indicates the percentage of transcripts which are associated to a particular GO biological process while the y-axis reports the significantly enriched GO classes.