Fig. 3.
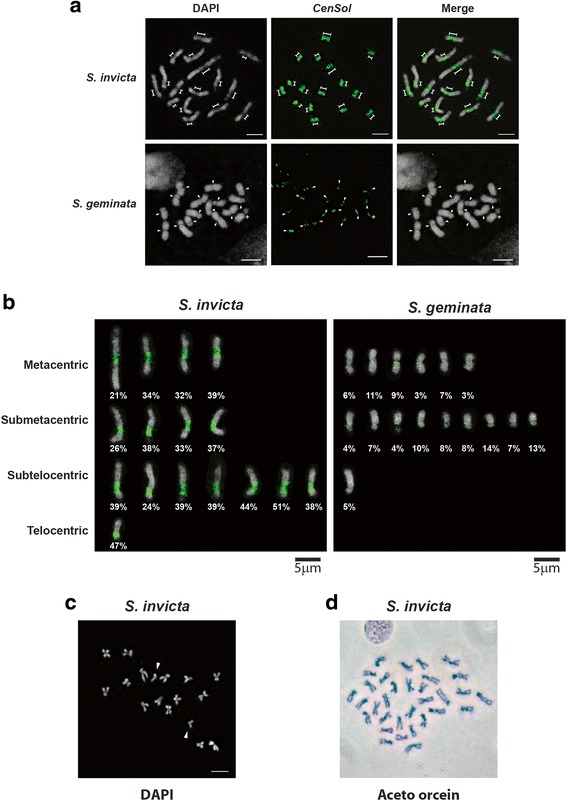
CenSol satellite fully localizes to the primary constriction of the haploid chromosomes in S. invicta and S. geminata. a and b FISH analysis with the CenSol probe (green); chromosomes counterstained with DAPI (gray). a The elongated (dimension lines) and the small (arrowheads) primary constrictions of chromosomes in S. invicta (upper panels) and S. geminata (lower panels) are indicated. b Positions of centromeres and CenSol coverage on chromosomes. Centromeric positions were categorized based on CenSol signal locations in FISH analysis. Chromosomes from S. invicta (left panel) and S. geminata (right panel) in (a) are artificially aligned and sorted according to the centromeric positions. The CenSol coverage rates (%) on each chromosome are labeled. c DAPI staining of the S. invicta chromosomes from a cell transitioning from metaphase to anaphase. Fifteen chromosome pairs are together while one pair has separated (arrowheads). d Aceto-orcein staining of metaphase chromosomes from male testes imaginal discs. Chromosome number is 32 likely because chromosomes come from two adjacent cells, although we cannot exclude potential diploidization which occurs in some cells of haploid male ants [58]. Scale bars, 5 μm
