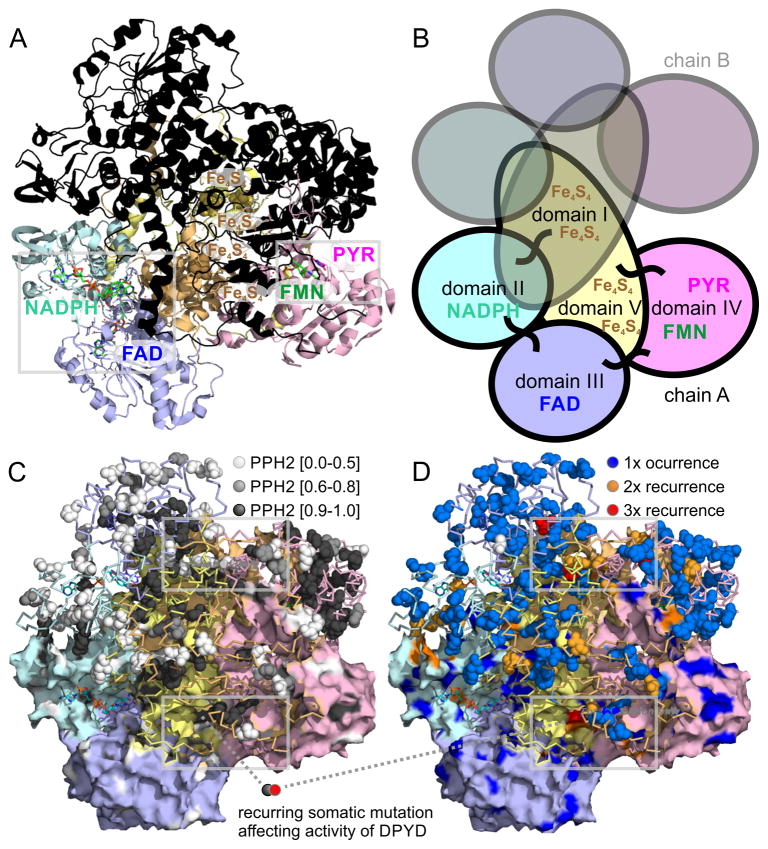
Figure 3. Structural analysis of DPYD dimer reveals hotspots of somatic mutations in ligand binding sites as well as interfaces of protein domains.
A) The 4Fe-4S cluster domain I of DPYD (uniprot entry Q12882 and PDB entry 1gth) is shown in orange, the FAD binding domain II in blue, the NADPH binding domain III in cyan, the pyrimidine and FMN binding domain IV in red, and the 4Fe-4S cluster domain V is shown in yellow. The lower monomer is color-coded according to domains and contains small-molecule ligands as sticks, the upper monomer is overlaid in black. B) The five domains of each DPYD monomer are color coded according to protein domains showing that the electron transfer chain crosses the dimer interface twice. The 4Fe-4S cluster domains I and V are intertwined and form extended inter-domain contacts. C) Functional impact of somatic missense mutations plotted as PPH2 scores onto surface (lower monomer; showing surface-bound mutations) and ribbon of DPYD structure (emphasizing accumulation of deleterious mutations in protein core in black). D) Recurrent missense somatic mutations in ligand binding sites as well as interfaces of protein domains of DPYD. Grey frames indicate regions of coincidence of high functional PPH2 scores, lack of surface accessibility coincides, and somatic recurrence shown enlarged in Figure 4.