Fig. 1.
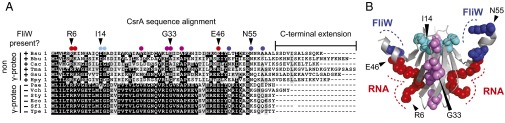
hag RNA and FliW interact with separate residues and surfaces of CsrA. (A) A multiple sequence alignment of CsrA primary sequences from γ-proteobacteria (“γ-proteo”) that do not encode FliW (−) and bacteria outside the γ-proteobacteria (“non γ-proteo”) that encode FliW (+). The following organisms were used to generate the multiple sequence alignment: B. subtilis 3610 (Bsu), Borrelia burgdorferi B31 (Bbu), Clostridium acetobutylicum ATCC824 (Cac), Thermotoga maritma MSB8 (Tma), Geobacter sulfurriducens PCA (Gsu), Helicobacter pylori J99 (Hpy), Pseudomonas aeruginosa PAO1 (Pae), Vibrio cholerae O1 El Tor (Vch), Salmonella typhimurium LT2 (Sty), E. coli MG1655 (Eco), Shigella flexneri 5 (Sfl), and Yersinia pestis KIM (Ype). Positions mutated in B. subtilis CsrA are indicated as colored circles above the multiple sequence alignment. Red circles indicate residues making up the RNA binding cleft, and purple circles indicate residues outside the RNA binding cleft that when mutated conferred a lof phenotype (38). Dark blue circles indicate poorly conserved residues with respect to γ-proteobacteria, and light blue circles indicate highly conserved residues that when mutated, conferred a CsrA resistant to FliW antagonism (crw) phenotype. (B) The side chain of each residue mutated by a lof allele (red or purple) or crw allele (dark blue or light blue) is shown and highlighted in the color corresponding to that used in panel 1A on the structure of the CsrA homolog RsmA from Pseudomonas aeruginosa (15). The RNA binding surface is indicated by a red dotted line. The location of residues R6, G33, and E46 required for CsrA function featured in the manuscript are indicated by carets. The predicted FliW binding surface is indicated by a blue dotted line. The location of residues I14 and N55 required for FliW antagonism featured in the manuscript are indicated by carets.