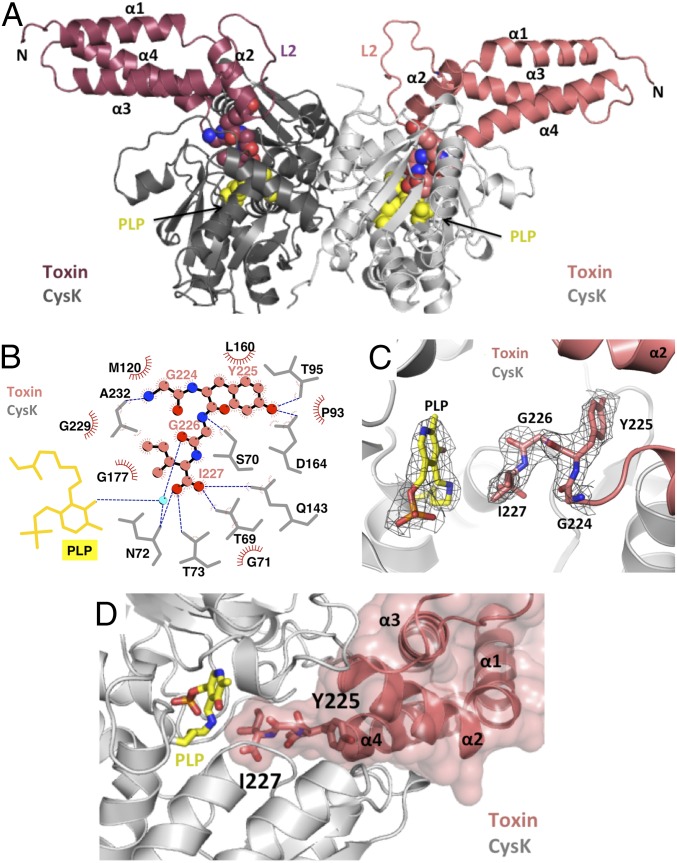
Fig. 1.
The CysK/CdiA-CTEC536 binary complex. (A) Crystal structure of the CysK/CdiA-CTEC536 complex. Secondary structure elements of the toxin nuclease domain are indicated together with flexible loop L2. The C-terminal GYGI peptides of CdiA-CTEC536 and CysK-bound pyridoxyl 5′-phosphate (PLP) are rendered as spheres. (B) GYGI peptide interaction network. CdiA-CTEC536 residues Gly224, Tyr225, Gly226, and Ile227 are shown in spheres, and CysK residues and PLP are rendered as gray and yellow sticks, respectively. A water molecule is shown as a cyan sphere. Red arcs represent hydrophobic interactions, and blue dashed lines indicate H-bonds. The interaction network was produced using LigPlot. (C) Interaction between CdiA-CTEC536 C-terminal GYGI peptide and the CysK active site. The CdiA-CTEC536 GYGI peptide and CysK PLP are shown in stick representation. The 2Fo − Fc electron density map of the CdiA-CTEC536 GYGI peptide and CysK PLP is shown in gray mesh and contoured at 1.0 σ. (D) The CdiA-CTEC536 toxin domain exploits shape complementarity to bind the CysK active-site cleft. Residues Gly224–Ile227 and PLP are shown in stick representation.