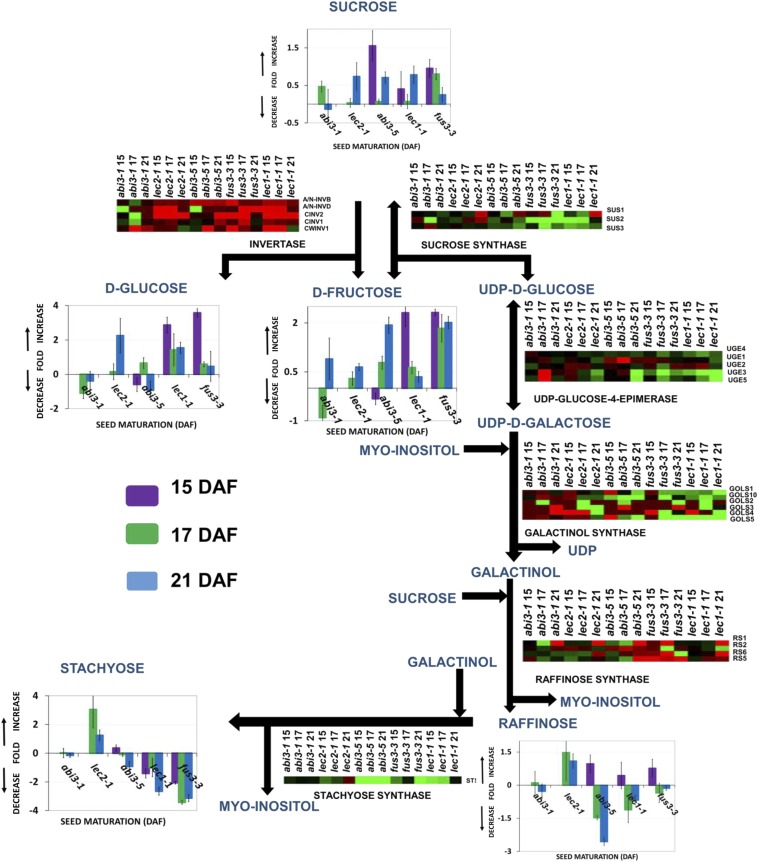
Fig. 2.
Metabolic pathway of stachyose biosynthesis in Arabidopsis seeds. Graphs represent changes in the contents of sugars and oligosaccharides for each mutant during the desiccation period. Bar graphs indicate the log2 fold change-relative content with respect to the wild-type controls. Values are from two independently grown sets of plants with three technical replicates each. Heat maps represent expression profiles of genes encoding putative enzymes involved in raffinose and stachyose synthesis as well as sucrose degradation pathways. Red and green represent up-regulated and down-regulated genes, respectively. The biochemical and physiological pathways were classified according to the KEGG database (www.genome.jp/kegg/). ALKALINE/NEUTRAL INVERTASE (A/N-INVB, AT4G34860; A/N-INVD, AT1G22650), CYTOSOLIC INVERTASE (CINV2, AT4G09510; CINV1, AT1G35580), CELL WALL INVERTASE 1 (CWINV1, AT3G13790), SUCROSE SYNTHASE (SUS1, AT5G20830; SUS2, AT5G49190; SUS3, AT4G02280), UDP-d-GLUCOSE/UDP-d-GALACTOSE 4-EPIMERASE (UGE1, AT1G12780; UGE2, AT4G23920; UGE3, AT1G63180; UGE4, AT1G64440; UGE5, AT4G10960), GALACTINOL SYNTHASE (GOLS1, AT2G47180; GOLS2, AT1G56600; GOLS3, AT1G09350; GOLS4, AT1G60470; GOLS5, AT5G23790; GOLS10, AT5G30500), RAFFINOSE SYNTHASE (RS1, AT1G55740; RS2, AT3G57520; RS5, AT5G40390; RS6, AT5G20250), STACHYOSE SYNTHASE (STS, AT4G01970). Data are the means ± SD of two biological replicates and three technical replicates.