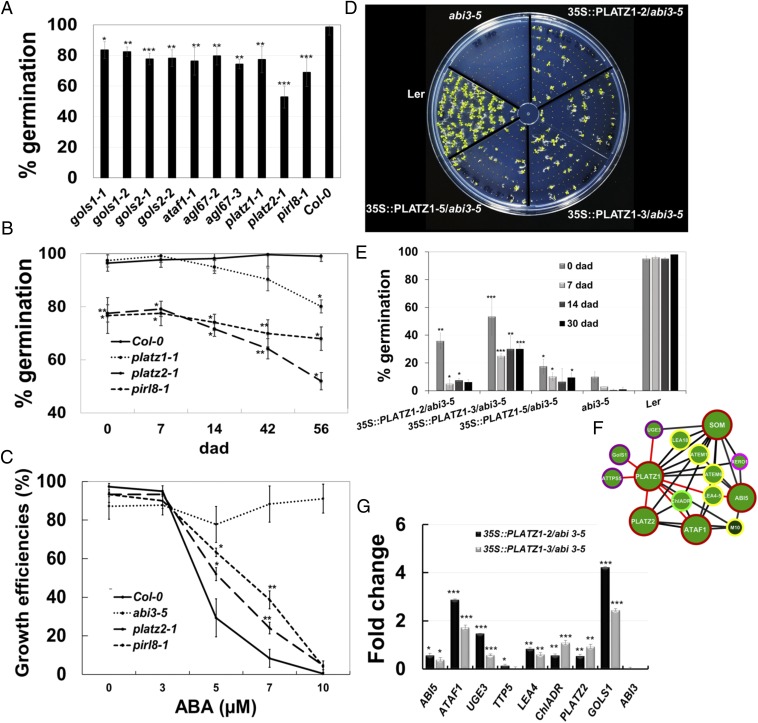
Fig. 4.
Transcriptomic network inference reveals novel genes involved in desiccation tolerance. (A) TF node mutants with reduced DT. Germination percentage of seeds 6 d after sowing. Values are the means ± SD of four biological replicates; n = 100 seeds per replicate. (B) Germination percentage of Col-0, platz1-1, platz2-1, and pirl8-1 seeds stored for 0, 7, 14, 43, and 56 d after desiccation (dad). Values are the means ± SD of three biological replicates. (C) Effect of ABA treatment on seedling viability or growth efficiency (scored as the % of seedlings with green cotyledons in each treatment); note that the highest percentage is detected for abi3-5 (reported as ABA-insensitive) and the lowest is for the WT control. Seedlings were grown on Murashige and Skoog (MS) medium with 0, 3, 5, 7, and 10 µM ABA and their phenotypes were scored 15 d after sowing. Values are the means ± SD of three replicates; n = 50 per replicate. (D) PLATZ1 overexpression partially rescues the abi3-5 seed desiccation intolerance phenotype. Seeds were grown on MS medium, and survival was scored 10 d after sowing. (E) Germination percentage of WT (Ler), abi3-5, and 35S::PLATZ1/abi3-5 seeds stored for 0, 7, 14, and 30 d after desiccation. Values are the means ± SD of three biological replicates. (F) PLATZ1 interactor genes (FullSeed-subNetDT2) related to cellular protection at DPI 0.1. (G) Real-time PCR of ABI5, ATAF1, UGE3, TTP5, LEA4, ChIADR, PLATZ2, GOLS1 (neighboring genes of PLATZ1 in F represented with red edges), and ABI3 in 35S::PLATZ1/abi3-5 line seeds at 21 DAF; abi3-5 was used as the reference gene. UBQ10 and TIP4L served as internal controls. Data are the means ± SD of two biological replicates and three technical replicates. Values are means ± SD of three biological replicates. Bars with asterisks are significantly different from the control (Student's t test, *P < 0.05, **P < 0.01, ***P < 0.0001).