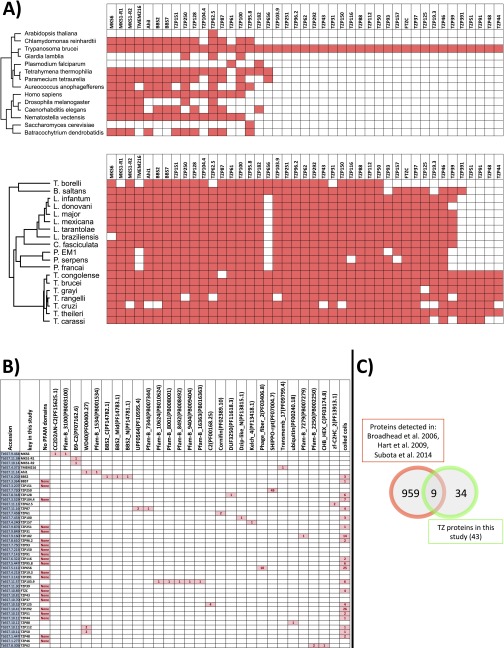
Fig. S2.
(A) An evolutionary tree of the proteins that were localized to the TZ in this study. Presence or absence in an evolutionary lineage was initially assessed using OrthoFinder (36) and refined using published literature, alignments, and hidden Markov models. Although many of the proteins are conserved, over half are specific to the kinetoplastids. Most TZPs are conserved within the kinetoplastids, but four TZPs are unique to the Trypanosoma spp. (B) Analysis of the PFAM domains and coiled coils of the TZPs. The B9-C2 PFAM was detected in B9-R1 and B9-R2. WD40 protein–protein interaction domain was detected in 3 different proteins, but is also found in 117 other predicted proteins in the trypanosome genome, suggesting that it does not have a function specifically associated with the TZ. Coiled coils were predicted using the program COILS (www.ch.embnet.org/software/COILS_form.html), with a window size of 21 residues and a cutoff score of 0.8, and it was revealed that some TZPs showed extensive coiled-coil structure, whereas other TZPs had no coiled coils predicted. (C) Most (>80%) of the TZPs identified in this study were not detected in any of the published trypanosome flagellum proteomes.