Abstract
Background
Malignant transformation of normal gastric cells is a complex and multistep process, resulting in development of heterogeneous tumours. Susceptible genetic background, accumulation of genetic changes, and environmental factors play an important role in gastric carcinogenesis. Single nucleotide polymorphisms (SNPs) in mitotic segregation genes could be responsible for inducing the slow process of accumulation of genetic changes, leading to genome instability.
Patients and methods
We performed a case-control study of polymorphisms in mitotic kinases TTK rs151658 and BUB1B rs1031963 and rs1801376 to assess their effects on gastric cancer risk. We examined the TTK abundance in gastric cancer tissues using immunoblot analysis.
Results
C/G genotype of rs151658 was more frequent in patients with diffuse type of gastric cancer and G/G genotype was more common in intestinal types of gastric cancers (p = 0.049). Polymorphic genotype A/A of rs1801376 was associated with higher risk for developing diffuse type of gastric cancer in female population (p = 0.007), whereas A/A frequencies were increased in male patients with subserosa tumour cell infiltration (p = 0.009). T/T genotype of rs1031963 was associated with well differentiated tumours (p = 0.035). TT+CT genotypes of rs1031963 and GG+AG genotypes of rs1801376 were significantly associated with gastric cancer risk (dominant model; OR = 2,929, 95% CI: 1.281-6.700; p = 0.017 and dominant model; OR = 0,364, 95% CI: 0.192-0.691; p = 0.003 respectively).
Conclusions
Our results suggest that polymorphisms in mitotic kinases TTK and BUB1B may contribute to gastric tumorigenesis and risk of tumour development. Further investigations on large populations and populations of different ethnicity are needed to determine their clinical utility.
Key words: cancer susceptibility, chromosomal instability, chromosome segregation, mitotic checkpoint, serine/threonine kinase, genetic association
Introduction
Gastric cancer is one of the major contributors to cancer-related deaths worldwide with estimated 989 600 new cases and 738 000 deaths in 2008.1,2 It is believed that complex interplay of genetic and environmental factors triggers the accumulation of numerous genetic and epigenetic alterations in cells, resulting in deregulation of normal cell functions and disruption of stomach linen homeostasis.3-6 Individual genetic factors probably contribute to aberrant processes in the genesis of malignant phenotype. Among them, single nucleotide polymorphisms (SNPs) and other genetic variants play an important role as the main genetic elements in the aetiology of several complex diseases, including gastric cancer.7-11 In gastric carcinogenesis this is further supported by the fact that only a small proportion of individuals exposed to the known environmental risk factors develop adenocarcinoma.5,10 Therefore, there is continuing interest for determining simple genetic tests for identifying individuals at high risk for the development of gastric tumours and for identifying patients with high risk for recurrence in order to ensure improved and early diagnosis as well as better survival of patients.
A majority of gastric cancer patients show chromosomal instability (CIN) resulting in aneuploidy.4,12,13 It has been suggested that tumour cells acquire aberrant chromosome numbers and other chromosomal defects as a result of deregulation of mechanisms responsible for maintaining the chromosomal number stability, such as spindle assembly checkpoint and chromosome segregation.14,15However, mutations in mitotic genes are rare, due to the fact that severe defects of these genes would trigger cell death by cell-surveillance early in the development.14-17 Studies revealed that subtle changes in mitotic segregation genes, controlling chromatids separation or regulating the progress of mitosis, could be prime candidates for inducing the slow process of accumulation of genetic changes, leading to CIN.15,18-20 The novel hypothesis is further supported by the fact that this process is slow, and explains the late onset of sporadic epithelial cancers21,22, as well as heterogeneous mutation load observed in different sections of tumours from individual patients.
The multidomain protein kinase BUB1B (BUB1-related kinase, known as MAD3 in yeast) plays a central role in the process of spindle assembly checkpoint (SAC), which prevents defects in the segregation of sister chromatids by delaying their separation until all chromatids have achieved correct attachments to the mitotic spindle.23,24 BUB1B is part of the mitotic checkpoint complex (MCC), which together with BUB3, MAD2 and CDC20 inhibits the anaphase-promoting complex/cyclosome (APC/C), delaying the onset of anaphase and ensuring proper chromosome segregation.25 The protein BUB1B has also been localized to the kinetochores and is important for stabilizing the kinetochore-microtubule interactions and chromosome alignment.26 A dual specificity protein kinase TTK (alias MPS1) is crucial for the spindle assembly checkpoint, for chromosome biorientation on the mitotic spindle and for ensuring accurate chromosome segregation.27,28 Inhibitor and chemical genetics studies showed that TTK activity facilitates the conformational activation of MAD2 from open to closed form (C-MAD2) capable of CDC20 binding and inhibition, thus delaying the onset of anaphase.29 TTK is probably implicated in the recruitment of the MAD1-C-MAD2 complex to kinetochores and during mitosis its activity is continuously required to recruit O-Mad2 to the Mad1-C-Mad2 core.30 Furthermore, TTK is required for CENP-E recruitment, whose activity is essential for metaphase chromosome alignment.30
In the present study we examined polymorphisms rs151658 (C>G) in TTK gene, rs1031963 (C>T) and rs1801376 (A>G) in BUB1B gene in the population of Slovenian patients with an advanced gastric cancer and their impact on gastric cancer risk. We also examined the associations of these genetic variants with clinico-histopathological features of patients.
Patients and methods
Research subjects
The study population (n = 284) consisted of 108 Slovenian patients with gastric cancer and 176 control subjects who at the time of peripheral blood extraction did not have cancer. Tumour and corresponding non-tumour tissues at least 7 cm away from the edge of the adenocarcinoma were collected from patients who were admitted to the Clinical Department for Abdominal Surgery at the University Medical Centre Ljubljana and Department for Pathology at the Institute of Oncology Ljubljana during the years 2000-2008. Samples were macrodissected by pathologist, frozen in liquid nitrogen and stored at −70°C. Comprehensive medical data were obtained from registries and pathologist’s evaluation. The following clinico-histopathological parameters were recorded: tumour differentiation (grade), location, blood and lymphatic vessel invasion (vascular invasion, perineural invasion), occurrence of tumour cells in the lymphatic vessels (lymphatic invasion), depth of invasion (pT), lymph node involvement (pN), and presence of distant metastases (pM). The gastric cancer cases were classified into diffuse type (n = 46) and intestinal (n = 58) according to Lauren classification. The mean age ± standard deviation (SD) of patients was 66.12 ± 12.02 (range, 33-87 years), and the percentage of men was 63.0%. Cases lost to follow-up (n = 6) and those, who died within 30 days after surgery (n = 2), were excluded from survival analyses. The control population was randomly selected during the years 1999-2007 and shared the ethnic and geographic background of the gastric cancer patients. The research was approved by the National Medical Ethics Committee of the Republic of Slovenia and confidentiality of personal medical data as well as other data relating to individual identification has been assured in accordance with the Helsinki Declaration.
Genotyping
Genomic DNA from gastric tumour and nontumour tissues was extracted using a Wizard® Genomic DNA Purification Kit (Promega, Madison, WI, USA) and QuickGene™ DNA Tissue Kit S (Fujifilm Corporation, Tokyo, Japan) on QuickGene-810 DNA isolation system (Fujifilm) according to manufacturer’s protocol. Genomic DNA from control population was extracted from peripheral blood samples using Wizard® Genomic DNA Purification Kit (Promega) following the manufacturer’s protocol. The DNA was quantified using a NanoDrop spectrophotometer (Thermo Fisher Scientific Inc.). Genotyping for polymorphism rs151658 (C>G) in TTK gene, and polymorphisms rs1031963 (C>T) and rs1801376 (A>G) in BUB1B gene was performed using TaqMan-based allele-specific polymerase chain reaction assays on the ABI Prism 7000 Sequencing Detection System apparatus (Applied Biosystems, Foster City, CA, USA) according to the procedure recommended by Applied Biosystems. The 10 μL reaction volume contained 100 ng of DNA. Assay IDs were: C_3181603_10, C_1237153_10, and C_3052718_1. In order to confirm the veracity of the results, the polymorphisms were re-genotyped by direct sequencing on a randomly selected smaller batch of samples.
Immunoblot analysis
A total of 21 paired gastric adenocarcinoma (GA) and adjacent control tissue samples were ground with a mortar and pestle in liquid nitrogen and lysed with 7 mol/L urea, 2 mol/L thiourea, 40 g/L CHAPS, with a protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO, USA). For every 10 mg tissue, 50 μl lysis buffer was added. After sonication on ice (3 × 10 s), the samples were incubated for 1 h on ice with occasional vortexing, and then centrifuged at 20,000 × g for 1 h at 4°C. The supernatants were collected and the protein concentrations were determined using the commercial Bradford reagent (Thermo Fisher Scientific, Waltham, MA, USA) with BSA used as the standard. Immunoblot analysis was performed on 42 samples. A total of 30 μg protein per sample was loaded onto 10% gels, separated using SDS-PAGE, and transferred onto PDVF membranes (Millipore, Billerica, MA, USA), which were then blocked in 50 g/L skimmed milk 1 h. The primary antibody was used in the following dilution: anti-Mpsl (anti-TTK) antibody, 1 pg/ml (ab11108, Abcam, Cambridge, UK). Horseradish peroxidase-conjugated secondary antibody was used in the following dilution: goat anti-mouse antibody, 1:5000 (115-035-062, Jackson ImmunoResearch, Newmarket, Suffolk, UK). The proteins were revealed by chemiluminescence using LAS-4000 CCD camera (Fujifilm, Tokyo, Japan). The blots were then quantified with Multi Gauge software (Fujifilm) and the intensities were normalized to Ponceau-S-stained membranes, to allow for loading and transfer variations.
Statistical and bioinformatic analyses
Statistical evaluation of the genotyping data was carried out using the χ2 or Fischer’s exact tests to compare the groups regarding genotype frequencies. Hardy-Weinberg (HW) equilibrium was calculated with an online program (http://www.genes.org.uk/software/hardy-weinberg.shtml).31 Survival was assessed by the Kaplan-Meier method and differences between groups were evaluated using the log-rank test. Multivariate survival analyses were further performed using the Cox proportional-hazards regression model. In the Cox multivariate analyses, forced entry procedure was used to determine the predictor variables. Only the variables that resulted in p-values < 0.05 in the Kaplan-Meier test were entered into the Cox proportional hazard model for the determination of independent prognostic factors for gastric cancer. The postoperative period was measured from the date of surgery to the date of the last follow-up or death. Statistical software used for calculations was IBM® SPSS® Statistics Version 20. For all statistical tests, a probability level (p-value) of less than 0.05 was considered significant.
To assess the statistical significance of altered protein abundance in the immunoblotting (as the tumour vs. non-tumour paired samples), non-parametric Wilcoxon signed-rank test was used. The tests were double-sided and the values with p < 0.05 with a confidence level of 95% were considered to be statistically significant. To assess the correlation of the altered protein abundance from the immunoblotting with the histopathological parameters, repeated measures ANOVA was used. The values with p < 0.05 were considered to be statistically significant. Bonferroni post-tests were used to determine where the differences were significant. All of the analyses were performed using Microsoft Office Excel 2007 (Microsoft Corporation, Washington, USA) and GraphPad Prism 5 (GraphPad Software, Inc., California, USA).
In order to functionally evaluate intronic polymorphisms we identified their effect in the context of polymorphic biological sequences on protein binding motifs. We used web-based software PROMO, which is part of the ALGGENE web-server.32,33 The search for putative binding sites was performed using the following parameters: human species, all motifs, and all factors. The data for comparisons of genotype frequencies in European populations of examined SNPs in this study was extracted from the 1000 Genomes Project data platform using a specific version of the Ensembl browser (http://browser.1000genomes.org).34
Results
Patients’ survival is associated with certain clinico-pathological features
The clinical information and demographic characteristics of selected patients with gastric cancer in this study are summarized in Table 1. At the end of a period of up to 11 years of follow-up, a total of 69 patients out of 100 have died.
Table 1.
Clinicopathological characteristics of patients with gastric cancer
| Parameter | Number of patients (%) |
|---|---|
| Age (years ± standard deviation) (n = 108) | 66.12 ± 12.02 |
| Gender (n = 105) | |
| Male | 68 (63.0) |
| Age (years ± standard deviation) (n = 66) | 65.07 ± 12.03 |
| Female | 40 (37.0) |
| Age (years ± standard deviation) (n = 39) | 67.90 ± 11.94 |
| Lauren’s classification (n = 104) | |
| Intestinal | 58 (55.8) |
| Diffuse | 46 (44.2) |
| Location (n = 101) | |
| Upper | 40 (39.6) |
| Lower | 34 (33.7) |
| Mixed | 27 (26.7) |
| Grade/differentiation (n = 105) | |
| Well | 9 (8.6) |
| Moderate | 24 (22.9) |
| Poor | 72 (68.6) |
| Vascular invasion (n = 80) | |
| Present | 27 (33.8) |
| Not present | 53 (66.3) |
| Perineural invasion (n = 95) | |
| Present | 44 (46.3) |
| Not present | 51 (53.7) |
| Lymphatic invasion (60) | |
| Present | 53 (88.3) |
| Not present | 7 (11.7) |
| pN (n = 105) | |
| 0 | 24 (22.9) |
| 1-2 | 15 (14.3) |
| 3-6 | 20 (19.0) |
| > 7 | 46 (43.8) |
| pT (n = 105) | |
| Muscularis propria | 6 (3.7) |
| Subserosa | 50 (42.6) |
| Serosa | 49 (36.1) |
pN = number of positive regional lymph nodes; pT = tumour invasion
The overall 5-year survival was 33.5%. No statistically significant association between tested genetic variations and survival was observed (p > 0.05). Univariate survival analysis showed that only Lauren’s classification and lymph node involvement (pN) were significant prognostic factors. Diffuse type predicted shorter 5-year survival (logrank test, χ2 = 5.516, p = 0.019) with overall mean estimate of survival for patients with intestinal type 64.67 months ± 7.74 (SE) (CI = 49.50-79.84) and 39.73 months ± 6.83 (SE) (CI = 26.75-53.11) for patients with diffuse type of gastric cancer. Regarding the parameter pN, patients with 7 or more positive lymph nodes had shorter survival time of 21.93 months ± 4.30 (SE) with CI = 13.5030-36 (log-rank test, χ2 = 34.169, p = 0.000). Multivariate analysis was performed for the same set of patients with complete clinical data sets. Cox regression model included both significant variables, pN and tumour classification. The enter method showed significant improvement (p < 0.05) if both parameters were entered into the model (Table 2).
Table 2.
Multivariate survival analysis of clinic-pathological variables in gastric cancer patients
| Variable | B* | SE (B) | OR (95% CI) | P |
|---|---|---|---|---|
| pN | 0.670 | 0.127 | 1.954 (1.525-2.504) | 0.000 |
| Lauren’s classification | 0.591 | 0.246 | 1.807 (1.116-2.925) | 0.016 |
Predicted change in the hazard for a unit increase in the predictor.
CI = confidence interval; OR = odds ratio; pN = number of positive regional lymph nodes; SE = standard error
SNPs in TTK and BUB1B are associated with type, grade, and location of gastric cancer
Associations between clinicopathological parameters and genotypes of SNPs are presented in Table 3. Statistical analysis revealed a weakly significant association for rs151658 genotypes C/G and risk of developing diffuse type of gastric cancer, and genotype G/G and risk of developing intestinal type of cancer (p = 0.049). Similar results were obtained for both male and female populations of patients (p = 0.047 and p = 0.024, respectively). Genotype A/A of rs1801376 polymorphism was significantly associated with higher risk of developing the diffuse type of gastric cancer in total and female populations of patients (p = 0.007). Interestingly, A/G genotype was under-represented in populations with diffuse type of gastric cancer. Genotype A/A of this polymorphism was also associated with the invasion of tumour cells into subserosa layer of stomach in male population (p = 0.009). A/A genotype was also associated with tumour location, namely, A/A frequencies were increased in patients with tumours disseminated across the whole stomach (p = 0.035).
Table 3.
Comparison of clinic-pathological features and genotypes TTK rs151658, BUB1B rs1031963, and BUB1B rs1801376 in patients with gastric cancer
| Parameter | Subject | Variant/Genotype | P | |||
|---|---|---|---|---|---|---|
| TTK rs151658 | ||||||
| GG | CG | CC | ||||
| Lauren’s classification | Intestinal | Total | 17 | 21 | 20 | 0.049 χ2=6.033 |
| Diffuse | 5 | 25 | 16 | |||
| Intestinal | Male | 9 | 10 | 14 | 0.047 χ2=6.113 | |
| Diffuse | 5 | 20 | 8 | |||
| Intestinal | Female | 8 | 11 | 6 | 0.024 F=7.499 | |
| Diffuse | 0 | 5 | 8 | |||
| BUB1B rs1801376 | ||||||
| AA | AG | GG | ||||
| Intestinal | Total | 18 | 33 | 7 | 0.007 | |
| Diffuse | 28 | 13 | 5 | χ2=9.951 | ||
| Intestinal | Male | 15 | 14 | 4 | 0.472 F=1.836 | |
| Diffuse | 20 | 9 | 4 | |||
| Intestinal | Female | 3 | 19 | 3 | 0.007 F=9.688 | |
| Diffuse | 8 | 4 | 1 | |||
| BUB1B rs1031963 | ||||||
| CC | CT | TT | ||||
| Tumour differentiation | Well | Total | 1 | 4 | 4 | |
| Moderate | 7 | 9 | 7 | 0.035 F=9.642 | ||
| Poor | 23 | 39 | 7 | |||
| Well | Male | 0 | 2 | 3 | ||
| Moderate | 6 | 7 | 2 | 0.139 F=6.439 | ||
| Poor | 19 | 20 | 6 | |||
| Well | Female | 1 | 2 | 1 | ||
| Moderate | 1 | 2 | 5 | 0.004 F=12.549 | ||
| Poor | 19 | 20 | 6 | |||
| BUB1B rs1801376 | ||||||
| AA | AG | GG | ||||
| pT | Muscularispropria | Total | 1 | 4 | 1 | 0.232 F=5.250 |
| Subserosa | 27 | 18 | 5 | |||
| Serosa | 18 | 25 | 6 | |||
| Muscularispropria | Male | 0 | 3 | 1 | 0.009 F=11.832 | |
| Subserosa | 22 | 6 | 2 | |||
| Serosa | 13 | 14 | 5 | |||
| Muscularispropria | Female | 1 | 1 | 0 | 0.816 F=1.967 | |
| Subserosa | 5 | 12 | 3 | |||
| Serosa | 5 | 11 | 1 | |||
| BUB1B rs1801376 | ||||||
| AA | AG | GG | ||||
| Tumour | Upper | Total | 14 | 23 | 3 | |
| location | Lower | 13 | 15 | 6 | 0.035 F=10.104 | |
| Whole | 18 | 6 | 3 | |||
pT = tumour invasion
Genotype T/T of rs1031963 was associated with well differentiated tumours in total population (p = 0.035); however, when we stratified it into female and male populations, we observed a significant association of this genotype with moderately differentiated tumours in the female population (p = 0.004). Clinico-pathological features lymph node involvement (pN), depth of invasion (pT), vascular invasion, perineural invasion and lymphatic invasion did not show significant associations with investigated polymorphisms.
SNPs in BUB1B are associated with gastric cancer risk
The analyses of genotype frequencies in selected SNPs between cases and controls are shown in Tables 4 and 5. The frequencies of all genotypes in cases and control groups were in Hardy-Weinberg equilibrium.
Table 4.
Distribution of genotype frequencies of TTK rs151658, BUB1B rs1031963, and BUB1B rs1801376 between gastric cancer patients and control subjects
| Variants | Genotype | Cases (n) | Controls (n) | p | HWE (cases) | HWE (controls) |
|---|---|---|---|---|---|---|
| TTK rs151658 | GG | 24 | 55 | χ2=3,628 0.163 | χ2=1.08 0.299 | χ2=2.87 0.090 |
| CG | 48 | 76 | ||||
| C/C | 36 | 44 | ||||
| BUB1B rs1031963 | CC | 32 | 48 | χ2=1.059 0.589 | χ2=0.345 0.557 | χ2=0.035 0.852 |
| CT | 54 | 89 | ||||
| TT | 18 | 39 | ||||
| BUB1B rs1801376 | AA | 47 | 89 | χ2=2.291 0.318 | χ2=0.021 0.885 | χ2=0.005 0.941 |
| AG | 49 | 69 | ||||
| GG | 12 | 13 | ||||
| Male population | AA | 36 | 49 | χ2=1.186 0.553 | χ2=1.530 0.216 | χ2=0.040 0.841 |
| AG | 24 | 39 | ||||
| GG | 8 | 7 | ||||
| Female population | AA | 11 | 40 | F=6.955 0.029 | χ2=3.352 0.067 | χ2=0.013 0.909 |
| AG | 25 | 30 | ||||
| GG | 4 | 6 |
F = Fisher statistics; HWE = Hardy-Weinberg Equilibrium; x2 = chi-square statistics;
Table 5.
Odds ratios for TTK rs151658, BUB1B rs1031963, and BUB1B rs1801376 between the cases and controls and their effect on gastric cancer risk
| Genotype model | Cases (n)/Control group (n) | OR (95% CI)* | P | PY |
|---|---|---|---|---|
| BUB1B rs1031963 | ||||
| Dominant | 72/120 | 0.900 | χ2=0.149 | χ2=0.062 |
| TT+CT vs. CC | vs. 32/48 | (0.527-1.536) | 0.699 | 0.803 |
| Recessive | 18/36 | 0.797 | χ2=0.507 | χ2=0.309 |
| TT vs. CT+CC | vs. 86/137 | (0.426-1.491) | 0.476 | 0.578 |
| Heterozygous | 54/89 | 0.910 | χ2=0.108 | χ2=0.035 |
| CT vs. CC | vs. 32/48 | (0.520-1.594) | 0.742 | 0.853 |
| Male population | ||||
| Dominant | 41/101 | 0.364 | χ2=9.848 | χ2=8.834 |
| TT+CT vs. CC | vs. 26/26 | (0.192-0.691) | 0.002 | 0.003 |
| Recessive | 11/26 | 0.763 | χ2=0.467 | χ2=0.241 |
| TT vs. CT+CC | vs. 56/101 | (0.351-1.659) | 0.494 | 0.623 |
| Heterozygous | 30/75 | 0.400 | χ2=6.959 | χ2=6.057 |
| CT vs. CC | vs. 26/26 | (0.201-0.797) | 0.008 | 0.014 |
| Female population | ||||
| Dominant | 31/57 | 1.994 | χ2=1.862 | χ2=1.281 |
| TT+CT vs. CC | vs. 6/22 | (0.731-5.437) | 0.172 | 0.258 |
| Recessive | 7/13 | 1.185 | χ2=0.107 | χ2=0.004 |
| TT vs. CT+CC | vs. 30/66 | (0.429-3.269) | 0.743 | 0.949 |
| Heterozygous | 24/44 | 2.000 | χ2=1.775 | χ2=1.188 |
| CT vs. CC | vs. 6/22 | (0.714-5.606) | 0.183 | 0.276 |
| BUB1B rs1801376 | ||||
| Dominant | 61/82 | 1.409 | χ2=1.927 | χ2=1.601 |
| GG+AG vs. AA | vs. 47/89 | (0.868-2.287) | 0.165 | 0.206 |
| Recessive | 12/13 | 1.519 | χ2=0.999 | χ2=0.615 |
| GG vs. AA+AG | vs. 96/158 | (0.666-3.465) | 0.318 | 0.433 |
| Heterozygous | 49/69 | 1.345 | χ2=1.304 | χ2=1.025 |
| AG vs. AA | vs. 47/89 | (0.808-2.237) | 0.253 | 0.311 |
| Malepopulation | ||||
| Dominant | 32/46 | 0.947 | χ2=0.029 | χ2=0.0002 |
| GG+AG vs. AA | vs. 36/49 | (0.508-1.766) | 0.864 | 0.990 |
| Recessive | 8/7 | 1.676 | χ2=0.917 | χ2=0.466 |
| GG vs. AA+AG | vs. 60/88 | (0.577-4.868) | 0.338 | 0.495 |
| Heterozygous | 24/39 | 0.838 | χ2=0.272 | χ2=0.124 |
| AG vs. AA | vs. 36/49 | (0.430-1.630) | 0.602 | 0.725 |
| Femalepopulation | ||||
| Dominant | 29/36 | 2.929 | χ2=6.719 | χ2=5.737 |
| GG+AG vs. AA | vs. 11/40 | (1.281-6.700) | 0.010 | 0.017 |
| Recessive | 4/6 | 1.296 | χ2=0.147 | χ2=0.001 |
| GG vs. AA+AG | vs. 36/70 | (0.344-4.889) | 0.701 | 0.971 |
| Heterozygous | 25/30 | 3.030 | χ2=6.732 | χ2=5.709 |
| AG vs. AA | vs. 11/40 | (1.292-7.108) | 0.009 | 0.017 |
p value with Yates correction
χ2 = chi-square statistics; OR = odds ratio; CI = confidence interval
The tested polymorphisms did not show significant differences between gastric cancer patients and control group. In contrast, when we stratified the population for gender, we found significant association between BUB1B rs1801376 genotypes and higher risk for developing gastric tumours (p = 0.029). Similarly, dominant model combining genotypes A/G and G/G showed comparable results (p = 0.010; p [Yates correction] = 0.017). Furthermore, tests for association showed analogous results and confirmed significantly higher frequency of G allele in female population of patients with gastric cancer (0.41 vs. 0.28 in control group). We also observed allele frequency difference in male patient population for BUB1B rs1031963. The dominant model, combining genotypes TT+CT versus CC, showed that patients with C/C homozygous allele had significantly higher risk for developing gastric cancer.
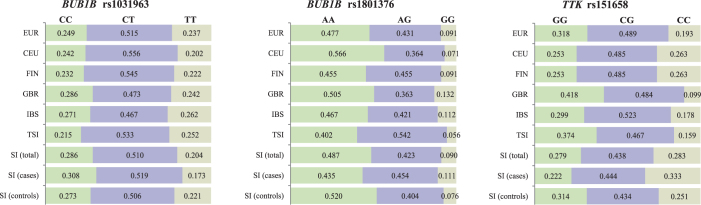
Distribution and genotype frequencies of SNPs in TKK and BUB1B between European and Slovenian populations
Comparisons of the SNPs’ genotype frequencies between our test groups and European populations are presented in Figure 3. Genotype frequencies of rs151658, rs1031963 and rs1801376 in our groups of populations showed significant differences from European population (Table 6). The frequency of rs151658 C/C genotype was higher than expected in the Slovenian population of patients compared to total European population (p = 0.015). Similarly, we observed more rs1031963 C/C genotypes in the male population of Slovenian patients with gastric cancer (p = 0.042) compared with total European population and European population stratified for males. The rs1801376 A/G genotype was higher and A/A genotype was under-represented in female population of patients with gastric cancer compared to the total European population (p = 0.034) and female European population (p = 0.014).
Figure 3.
Distribution of genotype frequencies of polymorphisms rs151658, rs1031963, and rs1801376 between European populations and Slovenian population.
CEU = Utah Residents (CEPH) with Northern and Western European Ancestry; EUR = European population; FIN = Finnish in Finland; GBR = British in England and Scotland; IBS = Iberian Population in Spain; SI (cases) = gastric cancer patients; SI (controls) = control population; SI (total) = combined populations of patients with gastric cancer and healthy controls; TSI = Tuscany in Italy
Table 6.
Comparison of TTK rs151658, BUB1B rs1031963, and BUB1B rs1801376 genotypes between the European population and examined groups of Slovenian population
| Population | N | Genotype counts | P |
|---|---|---|---|
| TTK rs151658 | |||
| EUR | 503 | 97 (C|C) / 246 (C|G) / 160 (G|G) | χ2 = 8.391; P = 0.015 a χ2 = 11.143; P = 0.004 b NS c |
| SI (total)a | 283 | 80 (C|C) / 124 (C|G) / 79 (G|G) | |
| SI (cases)b | 108 | 36 (C|C) / 48 (C|G) / 24 (G|G) | |
| SI (controls)c | 175 | 44 (C|C) / 76 (C|G) / 55 (G|G) | |
| BUB1B rs1031963 | |||
| EUR | 503 | 125 (C|C) / 259 (C|T) / 119 (T|T) | NS d NS e χ2 = 5.715; P = 0.057 f |
| EUR - male | 240 | 56 (C|C) / 124 (C|T) / 60 (T|T) | _ c _ d χ2 = 6.348; P = 0.042 f |
| SI (total)d | 280 | 80 (C|C) / 143 (C|T) / 57 (T|T) | |
| SI (cases - female)e | 36 | 6 (C|C) / 23 (C|T) / 7 (T|T) | |
| SI (cases - male)f | 65 | 25 (C|C) / 29 (C|T) / 11 (T|T) | |
| BUB1B rs1801376 | |||
| EUR | 503 | 240 (A|A) / 217 (A|G) / 46 (G|G) | NS g F = 6.569; P = 0.034 h NS 1 |
| EUR - female | 263 | 135 (A|A) / 109 (A|G) / 19 (G|G) | _ g F = 8.277; P = 0.014 h _ i |
| SI (total)g | 279 | 136 (A|A) / 118 (A|G) / 25 (G|G) | |
| SI (cases - female)h | 40 | 11 (A|A) / 25 (A|G) / 4 (G|G) | |
| SI (cases - male)i | 68 | 36 (A|A) / 24 (A|G) / 8 (G|G) |
EUR = European population; F = Fisher statistics; SI (cases) = gastric cancer patients; SI (controls) = control population; SI (total) = combined populations of patients and controls; χ2 = chi-square statistics; superscript letters indicate comparisons between European population and Slovenian populations
TTK abundance is altered in tumour tissues of gastric cancer patients
Immunobloting data on individual samples (Figure 1A) demonstrated statistical significance for the increased abundance of TTK (p = 0.03) in the tumour tissues. No statistically significant correlation of TTK abundance with clinical his topathological parameters or rs151658 genotypes was observed. However, some trends were observed (Figures 1B and C) for lymph node involvement (pN) and antral tumour location: TTK abundance was higher in normal tissues compared to tumour tissues when no regional nodes were invaded with tumour cells (pN = 0) and when the tumours were located at the bottom of the stomach (antrum).
Figure 1.
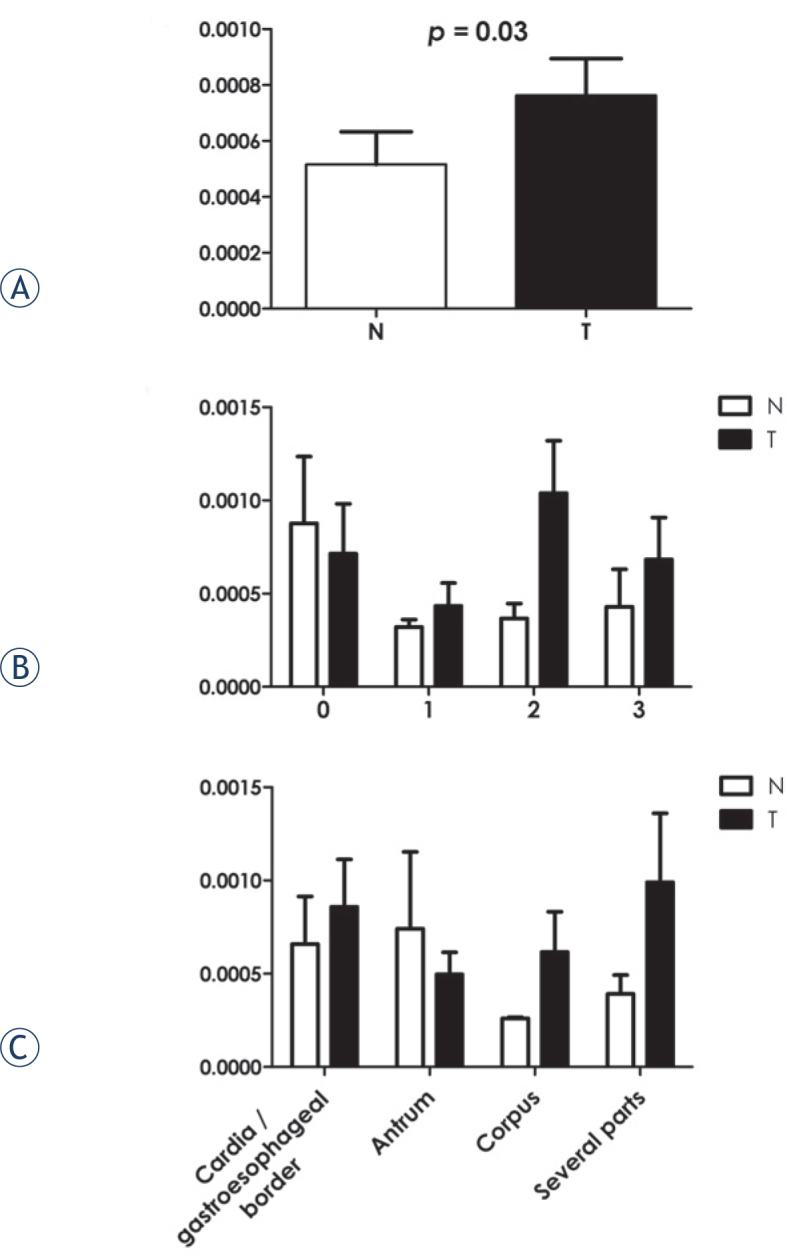
Immunoblotting of TTK. (A) Densitometry quantification analysis for the relative band densities from the protein abundance immunoblotting for the indicated protein in the non-tumour (N) and tumour (T) gastric tissues. The p value given (Wilcoxon signed-rank test) indicates the significance of the difference between the non-tumour (N) and tumour (T) gastric tissue samples. (B, C) Densitometry quantification analysis for the relative band densities from the protein abundance immunoblotting for TTK in the non-tumour (N) and tumour (T) gastric tissues samples according to lymph node involvement (pN) and location of the tumours.
Prediction of binding motifs showed that polymorphic sites in TTK and BUB1B bind different transcription factors
To determine if different intronic polymorphisms could affect binding of transcription factors, we performed in silico analysis of conserved human motifs using polymorphic sequences as templates (Figure 2). We identified distinct recognition sites for different proteins for both TTK rs151658 and BUB1B rs1031963. TFII-I, c-Myb, NFI/CTF, and HNF-4 alpha binding motifs were recognized if rs151658 polymorphic site contained C allele. In contrast, if G allele was present, GR, TFII-I, NCFI/CTF, and HNF-4 alpha were identified. Comparison of rs1031963 alleles showed that if allele T was present, several binding motifs were predicted, whereas in the case of allele C, there were no recognizable binding patterns.
Figure 2.
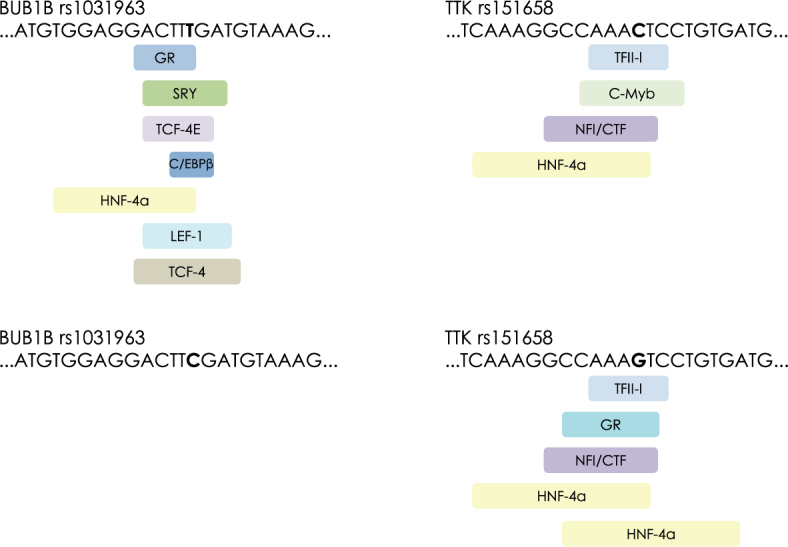
Predicted binding sites for transcription factors for polymorphic alleles rs1031963 and rs151658.
Discussion
In this study, we investigated the effects of selected polymorphisms in mitotic kinases TTK and BUB1Band risk of developing gastric cancer in Slovenian population. We also determined the associations between tested polymorphisms and clinic-pathological features of patients. The results provide evidence that TTK rs151658, BUB1B rs1031963, and rs1801376 could potentially serve as prognostic biomarkers for determining tumour differentiation and invasion. Furthermore, rs1801376 G allele could be used as one of determinants for gastric cancer screening in female population and CC genotype in rs1031963 could be used for selection of male population at higher risk for developing gastric cancer.
TTK gene harbours more than 600 different one-nucleotide polymorphisms (data obtained from GeneCards, http://www.genecards.org/). We investigated intronic polymorphism, because this gene has many alternative transcripts, and intronic one-nucleotide variants could have an effect on splicing and/or ubiquitination.35,36 SNP rs151658 lies between exons 12 and 13. In its vicinity there are binding sites for TFII-I, c-Myb, NFI/ CTF, and HNF-4 alpha transcription factors, if C allele is present and binding sites for GR, TFII-I, NFI/CTF, and HNF-4 alpha, if G allele is present (PROMO, ALGGEN server) (Figure 2).32,33 This indicated that different polymorphic alleles bind different proteins, which could in turn affect splicing or gene expression. Studies, performed on breast cancer patients, confirmed the significant association of this polymorphism with cancer risk15; however, we did not find any studies regarding the effect of rs151658 on gastric cancer risk. In our study, we identified the association of G/G geno-type with intestinal type of gastric cancer, while C/G genotype was significantly increased in cases with diffuse type of gastric cancer. Interestingly, comparison of genotype distribution for rs151658 between Slovenian patients with gastric cancer and European population showed that C/C genotype was over-represented in patients with gastric cancer. The significance of this finding is not clear and further analyses are needed on larger cohorts of patients in order to determine its usefulness in clinical setting.
To assess if the above mentioned genotypes perhaps had an effect on TTK protein levels, immunoblotting was performed. While the results regarding the effect of genotypes on protein abundance remain inconclusive, it should be noted that polymorphisms usually exert low-penetrance effects, which could more profoundly affect the pathogenesis of gastric cancer in early stages; however, when the disease progresses, the mutation load and aberrant expression of other genes mask their effects. We did, however, confirm higher abundance of TTK in tumour tissues, which is in accordance with several other studies and points out the deregulation of cell cycle homeostasis, higher proliferative trend of tumour cells and weakened spindle assembly checkpoint leading to increased genome instability and aneuploidy.37,38 Furthermore, this study showed a trend of increased TTK abundance associated with the spread of cancer cells to regional lymph nodes indicating a possible link between TTK levels and metastatic potential of malignant gastric cells.
Homozygous mutations of critical spindle-assembly BUB1B are extremely rare and associated with the diseases such as mosaic variegated aneuploidy syndrome 1 (biallelic mutations) and premature chromatid separation trait, which are both characterized by aneuploidy and chromosomal instability.39 BUB1B overexpression has been found in gastric cancers, although the results are often conflicting. In one study, the overexpression of BUB1B was associated with tumour proliferation40, however, Enjoji et al. observed that patients with higher expression of BUB1B had improved relapsefree survival.41 Furthermore, Ando et al. found that high expression of BUB1B correlated with invasion, lymph node metastasis, liver metastasis, and poor prognosis.14 Bohers et al. confirmed that the function of BUB1B is dosage-dependent by gradual reduction of BUB1B expression by shRNA in cell lines.42 In their experiment, residual levels of BUB1B protein below 50% of the normal level indicated premature chromatid separation and aneuploidy. These conflicting effects of BUB1B could be mediated by different polymorphisms, present in the nucleotide sequence of the gene. BUB1Brs1031963 polymorphism is in 5’-promoter region, which harbours binding sites for C/EBPbeta, GR, HNF-4alpha, LEF-1, SRY, TCF-4E, and TCF4 if T allele is present (PROMO, ALGGEN server).32,33Interestingly, if C allele is present, the DNA sequence harbours no transcription factor motifs. In our study the T/T genotype was associated with well differentiated tumours in total population, whereas in female population, when analysed separately, it was associated with moderately differentiated tumours. Well differentiated adenocarcinomas tend to have a better prognosis than infiltrative poorly differentiated adenocarcinomas. Furthermore, T/T+C/T genotypes were nominally associated with reduced risk of gastric cancer in male population, whereas C/C genotype was more common in male patient population. Comparisons with European population showed similar results. BUB1B rs1801376 A/A genotype was significantly higher in female patients with diffuse gastric cancer. A/A genotype was also increased in samples, which were characterized by invasion of tumour cells into subserosa in male population, and was associated with tumours, growing throughout whole stomach tissue. The consequence of this functional polymorphism is amino acid substitution Q349R in conserved region KEN, which is the binding site for CDC20.43 CDC20 is co-activator of anaphase promoting complex APC/C.24 Impaired function of KEN region in BUB1B could thus affect the regulation of anaphase delay, which ensures genome stability by providing time for correct spindle assembly, chromosome alignment and segregation. In addition, A/G genotype showed significant association with gastric cancer risk in female population of gastric cancer patients compared to Slovenian control group and European population.
In conclusion, our study provides evidence that polymorphisms in mitotic kinases TTK rs151658, BUB1B rs1031963 and rs1801376 could have an effect on gastric tumorigenesis and risk of adenocarcinoma development. In addition, we observed differences in genotype distributions between certain clinic-pathological features in patient populations, which could be used as the diagnostic aid in clinical setting; however, a large scale evaluation of these polymorphisms and functional analyses of their effect on protein products are needed to confirm their role in gastric carcinogenesis.
Disclosure: No potential conflicts of interest were disclosed.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Winder T, Wilson PM, Yang D, Zhang W, Ning Y, Power DG. et al. An individual coding polymorphism and the haplotype of the SPARC gene predict gastric cancer recurrence. Pharmacogenomics J. 2013;13:342–348. doi: 10.1038/tpj.2012.11. [DOI] [PubMed] [Google Scholar]
- 3.Hudler P, Repse S, Juvan R, Komel R. A genomic approach to investigate expression profiles in Slovenian patients with gastric cancer. Oncol Lett. 2011;2:1003–1004. doi: 10.3892/ol.2011.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ottini L, Falchetti M, Lupi R, Rizzolo P, Agnese V, Colucci G. et al. Patterns of genomic instability in gastric cancer: clinical implications and perspectives. Ann Oncol. 2006;17:vii97–vii102. doi: 10.1093/annonc/mdl960. [DOI] [PubMed] [Google Scholar]
- 5.Resende C, Ristimaki A, Machado JC. Genetic and epigenetic alteration in gastric carcinogenesis. Helicobacter. 2010;15(Suppl 1):34–39. doi: 10.1111/j.1523-5378.2010.00782.x. [DOI] [PubMed] [Google Scholar]
- 6.Tahara E. Abnormal growth factor/cytokine network in gastric cancer. Cancer Microenviron. 2008;1:85–91. doi: 10.1007/s12307-008-0008-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen J, Liu NN, Li JQ, Yang L, Zeng Y, Zhao XM. et al. Association between ITGA2 C807T polymorphism and gastric cancer risk. World J Gastroenterol. 2011;17:2860–2866. doi: 10.3748/wjg.v17.i23.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gonzalez CA, Agudo A. Carcinogenesis, prevention and early detection of gastric cancer: where we are and where we should go. Int J Cancer. 2012;130:745–753. doi: 10.1002/ijc.26430. [DOI] [PubMed] [Google Scholar]
- 9.Milne AN, Carneiro F, O’morain C, Offerhaus GJ. Nature meets nurture: molecular genetics of gastric cancer. Hum Genet. 2009;126:615–628. doi: 10.1007/s00439-009-0722-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pinheiro DdR, Ferreira WA, Barros MB, Araujo MD, Rodrigues-antunes S, Borges bdo N. Perspectives on new biomarkers in gastric cancer: diagnostic and prognostic applications. World J Gastroenterol. 2014;20:11574–11585. doi: 10.3748/wjg.v20.i33.11574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan P. Germline polymorphisms as modulators of cancer phenotypes. BMC Med. 2008;6:27. doi: 10.1186/1741-7015-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bass AJ, Thorsson V, Shmulevich I, Reynolds SM, Miller M, Bernard B. et al. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grabsch HI, Askham JM, Morrison EE, Pomjanski N, Lickvers K, Parsons WJ. et al. Expression of BUB1 protein in gastric cancer correlates with the histological subtype, but not with DNA ploidy or microsatellite instability. J Pathol. 2004;202:208–214. doi: 10.1002/path.1499. [DOI] [PubMed] [Google Scholar]
- 14.Ando K, Kakeji Y, Kitao H, Iimori M, Zhao Y, Yoshida R. et al. High expression of BUBR1 is one of the factors for inducing DNA aneuploidy and progression in gastric cancer. Cancer Sci. 2010;101:639–645. doi: 10.1111/j.1349-7006.2009.01457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lo YL, Yu JC, Chen ST, Hsu GC, Mau YC, Yang SL. et al. Breast cancer risk associated with genotypic polymorphism of the mitotic checkpoint genes: a multigenic study on cancer susceptibility. Carcinogenesis. 2007;28:1079–1086. doi: 10.1093/carcin/bgl256. [DOI] [PubMed] [Google Scholar]
- 16.Castro dIP, Carcer dG, Malumbres M. A census of mitotic cancer genes: New insights into tumor cell biology and cancer therapy. Carcinogenesis. 2007;28:899–912. doi: 10.1093/carcin/bgm019. [DOI] [PubMed] [Google Scholar]
- 17.Yuan B, Xu Y, Woo JH, Wang Y, Bae YK, Yoon DS. et al. Increased expression of mitotic checkpoint genes in breast cancer cells with chromosomal instability. Clin Cancer Res. 2006;12:405–410. doi: 10.1158/1078-0432.CCR-05-0903. [DOI] [PubMed] [Google Scholar]
- 18.Couch FJ, Wang X, Bamlet WR, de andrade M, Petersen GM, McWilliams RR. Association of mitotic regulation pathway polymorphisms with pancreatic cancer risk and outcome. Cancer Epidemiol Biomarkers Prev. 2010;19:251–257. doi: 10.1158/1055-9965.EPI-09-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang P, Wang Y, Yan H, Xie Q, Zhao L, Xu S. et al. Genetic variation in the major mitotic checkpoint genes and risk of breast cancer: a multigenic study on cancer susceptibility. Tumour Biol. 2014;35:6701–6705. doi: 10.1007/s13277-014-1906-0. [DOI] [PubMed] [Google Scholar]
- 20.Wang X, Fredericksen ZS, Vierkant RA, Kosel ML, Pankratz VS, Cerhan JR. et al. Association of genetic variation in mitotic kinases with breast cancer risk. Breast Cancer Res Treat. 2010;119:453–462. doi: 10.1007/s10549-009-0404-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duesberg P, Rasnick D. Aneuploidy, the somatic mutation that makes cancer a species of its own. CellMotil Cytoskeleton. 2000;47:81–107. doi: 10.1002/1097-0169(200010)47:2<81::AID-CM1>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 22.Frank SA. Genetic predisposition to cancer - insights from population genetics. Nat Rev Genet. 2004;5:764–772. doi: 10.1038/nrg1450. [DOI] [PubMed] [Google Scholar]
- 23.Bolanos-garcia VM, Lischetti T, Matak-vinkovic D, Cota E, Simpson PJ, Chirgadze DY. et al. Structure of a Blinkin-BUBR1 complex reveals an interaction crucial for kinetochore-mitotic checkpoint regulation via an unanticipated binding site. Structure. 2011;19:1691–1700. doi: 10.1016/j.str.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chao WC, Kulkarni K, Zhang Z, Kong EH, Barford D. Structure of the mitotic checkpoint complex. Nature. 2012;484:208–213. doi: 10.1038/nature10896. [DOI] [PubMed] [Google Scholar]
- 25.Tian W, Li B, Warrington R, Tomchick DR, Yu H, Luo X. Structural analysis of human Cdc20 supports multisite degron recognition by APC/C. Proc Natl Acad Sci U S A. 2012;109:18419–184124. doi: 10.1073/pnas.1213438109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bolanos-garcia VM, Blundell TL. BUB1 and BUBR1: multifaceted kinases of the cell cycle. Trends Biochem Sci. 2011;36:141–150. doi: 10.1016/j.tibs.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kagami Y, Nihira K, Wada S, Ono M, Honda M, Yoshida K. Mps1 phosphorylation of condensin II controls chromosome condensation at the onset of mitosis. J Cell Biol. 2014;205:781–790. doi: 10.1083/jcb.201308172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.antaguida S, Tighe A, D’alise AM, Taylor SS, Musacchio A. Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine. J Cell Biol. 2010;190:73–87. doi: 10.1083/jcb.201001036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee S, Thebault P, Freschi L, Beaufils S, Blundell TL, Landry CR. et al. Characterization of spindle checkpoint kinase Mps1 reveals domain with functional and structural similarities to tetratricopeptide repeat motifs of Bub1 and BubR1 checkpoint kinases. J Biol Chem. 2012;287:5988–6001. doi: 10.1074/jbc.M111.307355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hewitt L, Tighe A, Santaguida S, White AM, Jones CD, Musacchio A. et al. Sustained Mps1 activity is required in mitosis to recruit O-Mad2 to the Mad1-C-Mad2 core complex. J Cell Biol. 2010;190:25–34. doi: 10.1083/jcb.201002133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodriguez S, Gaunt TR, Day IN. Hardy-Weinberg equilibrium testing of biological ascertainment for Mendelian randomization studies. Am J Epidemiol. 2009;169:505–514. doi: 10.1093/aje/kwn359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Farre D, Roset R, Huerta M, Adsuara JE, Rosello L, Alba MM. et al. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 2003;31:3651–3653. doi: 10.1093/nar/gkg605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Messeguer X, Escudero R, Farre D, Nunez O, Martinez J, Alba MM. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics. 2002;18:333–334. doi: 10.1093/bioinformatics/18.2.333. [DOI] [PubMed] [Google Scholar]
- 34.Abecasis GR, Auton A, Brooks LD, Depristo MA, Durbin RM, Handsaker RE. et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cui Y, Cheng X, Zhang C, Zhang Y, Li S, Wang C. et al. Degradation of the human mitotic checkpoint kinase Mps1 is cell cycle-regulated by APC-cCdc20 and APC-cCdh1 ubiquitin ligases. J Biol Chem. 2010;285:32988–32998. doi: 10.1074/jbc.M110.140905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun T, Yang X, Wang W, Zhang X, Xu Q, Zhu S. et al. Cellular abundance of Mps1 and the role of its carboxyl terminal tail in substrate recruitment. J Biol Chem. 2010;285:38730–38739. doi: 10.1074/jbc.M110.177642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaistha BP, Honstein T, Muller V, Bielak S, Sauer M, Kreider R. et al. Key role of dual specificity kinase TTK in proliferation and survival of pancreatic cancer cells. Br J Cancer. 2014;111:1780–1787. doi: 10.1038/bjc.2014.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ling Y, Zhang X, Bai Y, Li P, Wei C, Song T. et al. Overexpression of Mps1 in colon cancer cells attenuates the spindle assembly checkpoint and increases aneuploidy. Biochem Biophys Res Commun. 2014;450:1690–1695. doi: 10.1016/j.bbrc.2014.07.071. [DOI] [PubMed] [Google Scholar]
- 39.Rio frio T, Lavoie J, Hamel N, Geyer FC, Kushner YB, Novak DJ. et al. Homozygous BUB1B mutation and susceptibility to gastrointestinal neoplasia. N Engl J Med. 2010;363:2628–2637. doi: 10.1056/NEJMoa1006565. [DOI] [PubMed] [Google Scholar]
- 40.Grabsch H, Takeno S, Parsons WJ, Pomjanski N, Boecking A, Gabbert HE. et al. Overexpression of the mitotic checkpoint genes BUB1, BUBR1, and BUB3 in gastric cancer--association with tumour cell proliferation. J Pathol. 2003;200:16–22. doi: 10.1002/path.1324. [DOI] [PubMed] [Google Scholar]
- 41.Enjoji M, lida S, Sugita H, Ishikawa T, Uetake H, Inokuchi M. et al. BubR1 and AURKB overexpression are associated with a favorable prognosis in gastric cancer. Mol Med Report. 2009;2:589–596. doi: 10.3892/mmr_00000142. [DOI] [PubMed] [Google Scholar]
- 42.Bohers E, Sarafan-vasseur N, Drouet A, Paresy M, Latouche JB, Flaman JM. et al. Gradual reduction of BUBR1 protein levels results in premature sister-chromatid separation then in aneuploidy. Hum Genet. 2008;124:473–478. doi: 10.1007/s00439-008-0572-y. [DOI] [PubMed] [Google Scholar]
- 43.D’arcy S, Davies OR, Blundell TL, Bolanos-garcia VM. Defining the molecular basis of BubR1 kinetochore interactions and APC/C-CDC20 inhibition. J Biol Chem. 2010;285:14764–14776. doi: 10.1074/jbc.M109.082016. [DOI] [PMC free article] [PubMed] [Google Scholar]