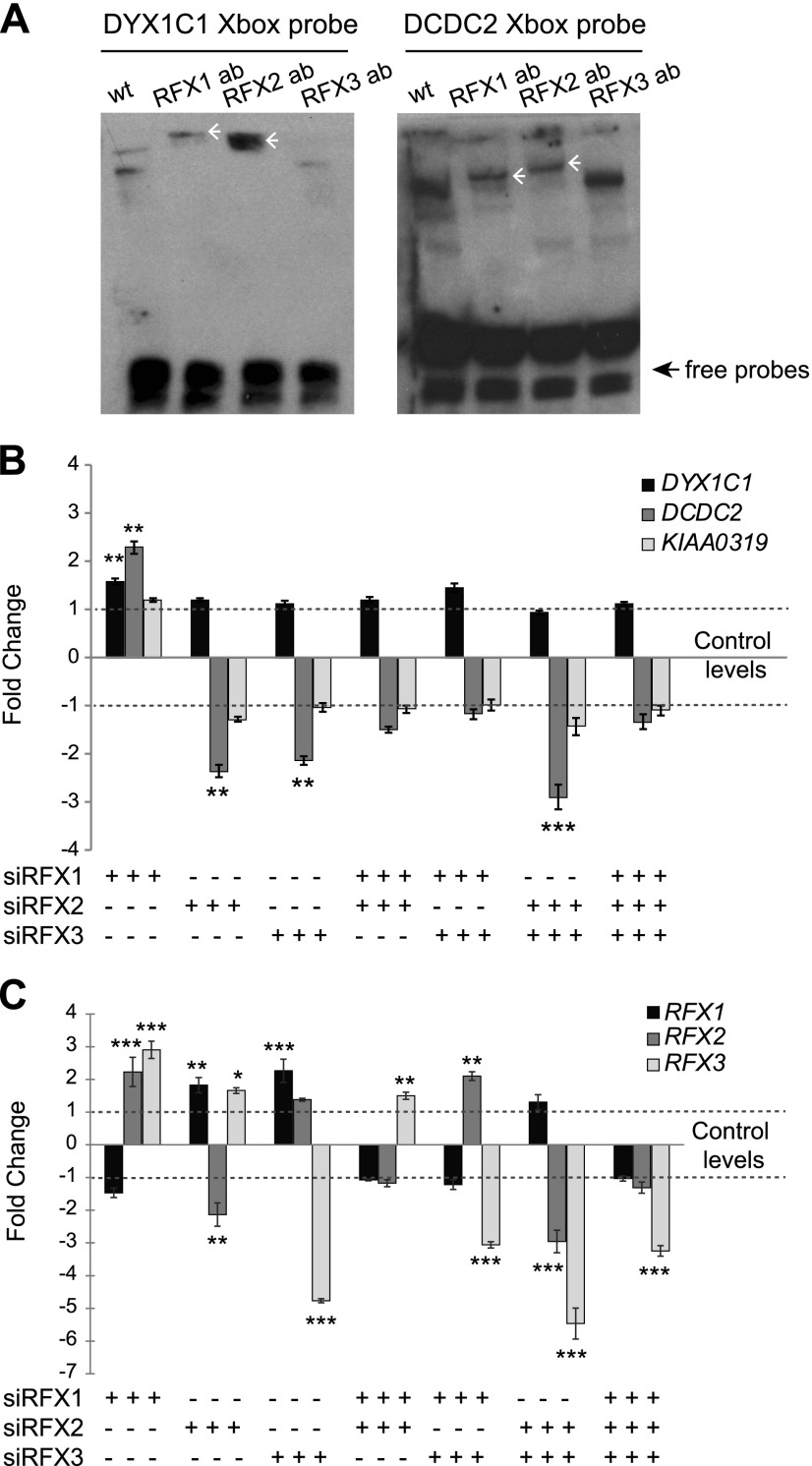
Figure 2.
Knockdown of RFX1, RFX2, and RFX3 affect the expression of DYX1C1 and DCDC2 but not KIAA0319 in hTERT-RPE1 cells. A) Binding of RFX1 and RFX2 to the X-box motifs present in DYX1C1 and DCDC2 promoters. Biotinylated probes spanning the X-box motifs were incubated with nuclear extracts from serum-starved hTERT-RPE1 cells with or without antibodies (ab) against RFX1, RFX2, and RFX3. Supershifts for both probes were detected for RFX1 and RFX2 antibodies (white arrowheads). One representative experiment of 3 is shown. B) Fold-change differences in the expression of DCGs upon knockdown of RFX TFs. By using siRNA against RFX1, RFX2, and RFX3, alone or in different combinations, genes were silenced in hTERT-RPE1 cells. Cells were thereafter starved for 24 h to induce ciliogenesis, and expression levels of DYX1C1, DCDC2, and KIAA0319 were measured by using qRT-PCR. C) Fold-change difference in expression levels of RFX1, RFX2, and RFX3 upon siRNA silencing. qRT-PCR data are summarized by using the ΔΔCt method as displayed as mean fold-change (2−ΔΔCt or −2ΔΔCt) ± sem. Significance of the post hoc tests after ANOVA is presented as ***P ≤ 0.001, **P ≤ 0.01, *P ≤ 0.05 after multiple correction of simultaneous comparisons.