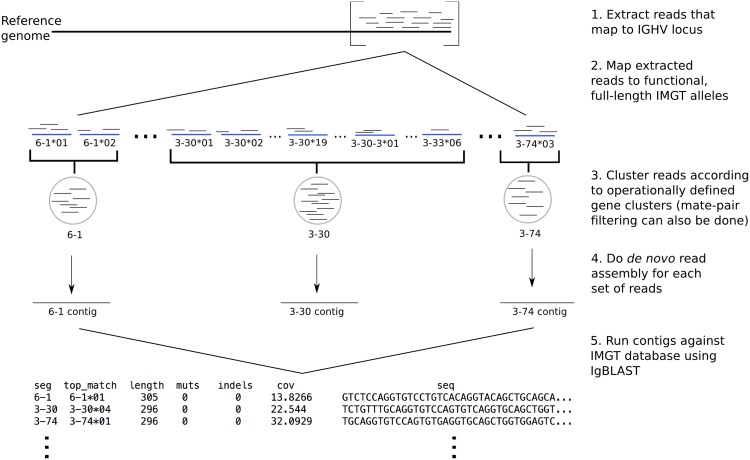
Fig 2. Schematic of genotyping pipeline.
1. WGS reads (short thin black horizontal lines) that map to the IGHV locus of a single individual are extracted from full set of reads. 2. These extracted reads are mapped using Bowtie2 [36] to known functional V gene segment alleles (thick blue horizontal lines) curated from the IMGT database [37]. 3. Mapped reads are pooled according to the gene clusters described in the Results section. At this stage, extra filtering steps using additional information like mate-pair data can also be applied. 4. Local assembly using SPAdes [38] is performed on reads to produce contigs (long thin black horizontal lines) corresponding to each gene cluster. 5. The resulting contigs are identified using stand-alone IgBLAST [39]. The final output contains, for each individual and each assembled contig: the closest-matching existing allele, the length of match, the number of nucleotide mutations or indels that separate the contig from the closest-matching allele, the read coverage of the contig as reported by SPAdes, and the nucleotide sequence of the contig.