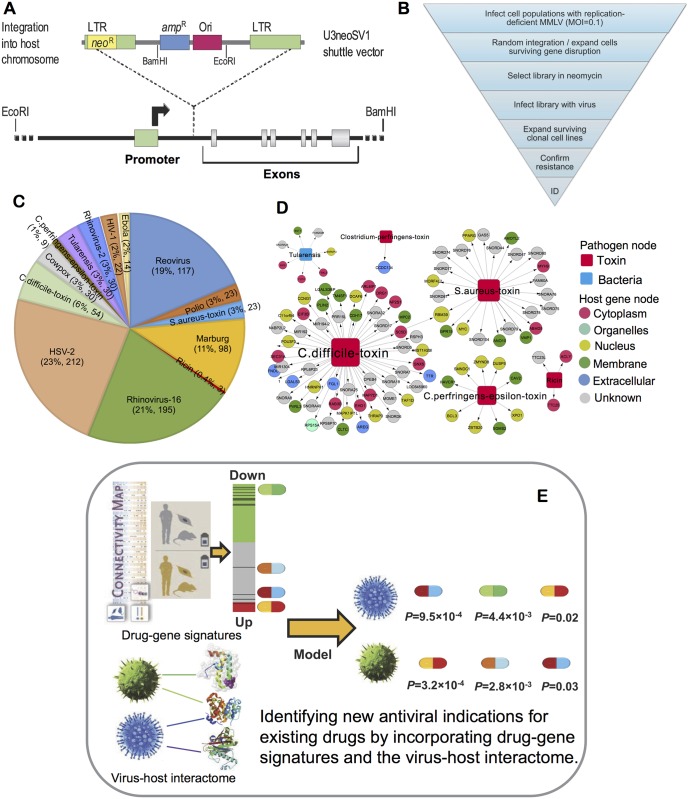
Fig 1. Diagram of the integrative antiviral drug discovery pipeline.
(A) The gene-trap insertional mutagenesis approach employs an MMLV-based shuttle vector that randomly integrates into host cell chromosomes and contains a promoterless neomycin-resistance gene. Shuttle vector integration between a host-cell promoter and an early exon disrupts (traps) the gene, allowing neomycin selection and derivation of a gene-trap library. (B) Host genes mediating the toxic effects of lytic viral replication or exposure to toxins were identified by: (i) selecting gene-trap libraries in neomycin; (ii) exposing gene-trap library cells to a lytic virus or a toxin; (iii) isolating surviving clones; (iv) resistance confirmation in surviving clones following exposure to a 10-fold higher dose of the virus or toxin studied; and (v) identification of the trapped gene by digesting genomic DNA to liberate shuttle vectors, self-ligation, bacterial transform, ampicillin selection, and sequencing trapped genes in the recovered plasmids. (C) Distribution of newly discovered virus-host interaction pairs for 10 viruses, 1 bacterium, and 5 toxins. (D) Global pathogen-host interaction network identified by genome-wide gene-trap insertional mutagenesis, where toxins and bacteria are represented by red and cyan squares respectively. The host cell gene products (circles) are colored based on their subcellular locations collected from the LocDB (https://www.rostlab.org/services/locDB/). (E) Identification of candidates for antiviral drug repositioning approach by incorporating drug-gene signatures from the Connectivity Map into the global virus-host interactome.