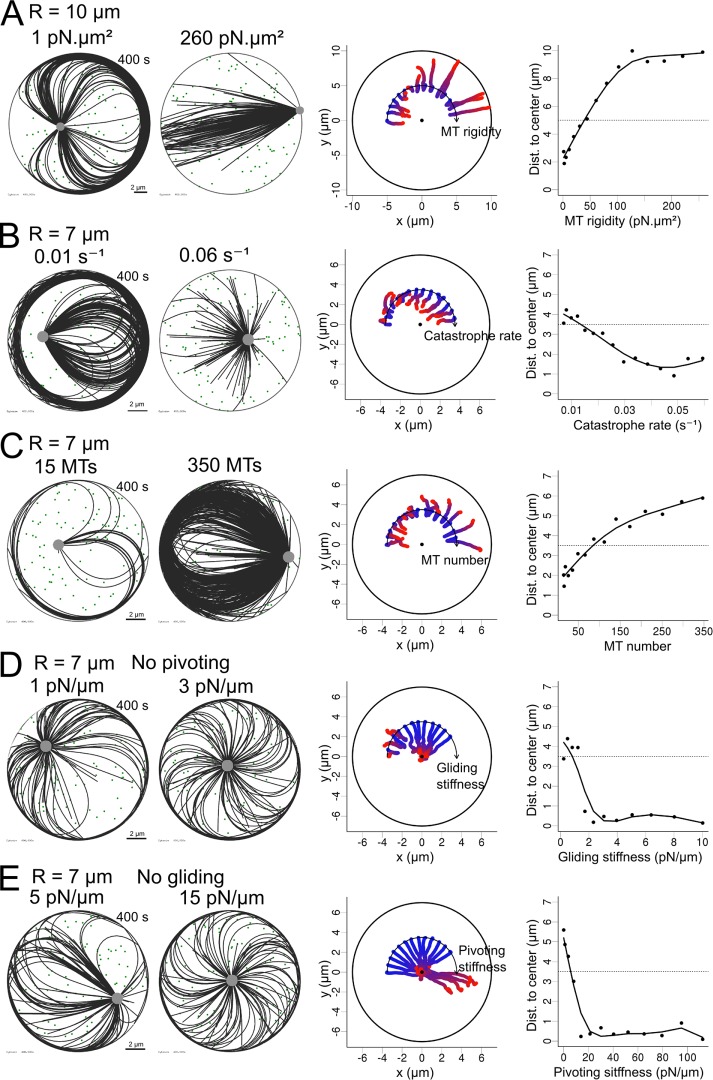
FIGURE 5:
Transitions between centering and decentering regimes. (A–E) Simulations in which one parameter was geometrically varied: MT rigidity, from 1 to 260 pN/μm2; MT unloaded catastrophe rate, from 0.01–0.06 s−1; number of MTs, from 15 to 350; MT gliding stiffness while pivoting is not allowed, from 0 to 10 pN/μm; and MT pivoting stiffness when gliding is not allowed, from 0 to 110 pN/μm. In each case, 100 cytoplasmic dynein motors were randomly positioned in the cell and one parameter of the system was varied systematically. Cell radius is 10 μm in A and 7 μm in B–E. Left, exemplary simulations for two different outcomes observed while varying a parameter. Middle, trajectories of simulations obtained while varying a parameter, displayed along an arc, with values increasing from left to right. For each trajectory, time is indicated by the color, from blue (0 s) to red (400 s). The center of each disk is marked with a black dot. Right, centrosome positioning as a function of various parameters, measured by the distance to the cell center. The dotted line represents the initial centrosome–center distance (half of the confinement radius). In A–C, MTs were allowed to pivot and glide.