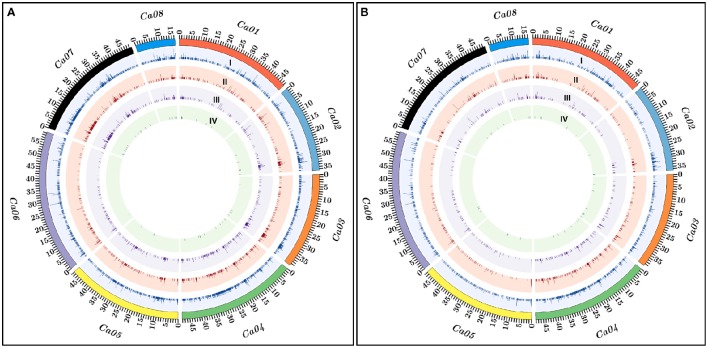
Figure 2.
The relative genomic distribution of PI (Pusa 1103 × ILWC 46) (A) and PII (Pusa 256 × ILWC 46) (B) mapping populations-derived polymorphic InDel markers physically mapped on eight chromosomes of kabuli chickpea genome are depicted by the Circos circular ideograms. The outermost circles represent the different physical sizes (Mb) of eight chromosomes coded with multiple colors as per the pseudomolecule sizes documented in kabuli chickpea genome (Varshney et al., 2013). Total InDel markers (I) including gene-derived (II), regulatory (III), and coding (IV) markers polymorphic between high and low pod number parental accessions and homozygous bulks of two inter-specific mapping populations—PI (Pusa 1103 × ILWC 46) (A) and PII (Pusa 256 × ILWC 46) (B) are indicated.