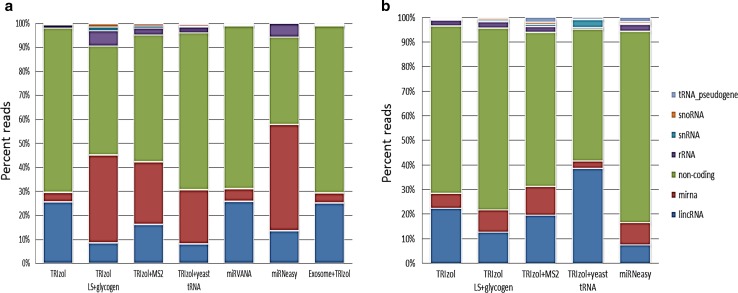
Fig. 7.
Reads aligned to other RNA species: The processed reads were aligned to the Ensembl database and the percent of reads aligning to different RNA species is shown. a Unpooled samples b Pooled samples. Chi square comparisons for small RNA species were significant at p value; considering the variability within methods, Kruskal–Wallis rank sum analysis was performed. Unpooled samples had Chi squared = 99.021, df = 11, p-value = 2.79e–16 and pooled samples had Chi squared = 191.56, df = 11, p-value < 2.2e–16. The p-value turns out to be nearly zero and thus rejecting null hypothesis and small RNA species population are not identical in different methods