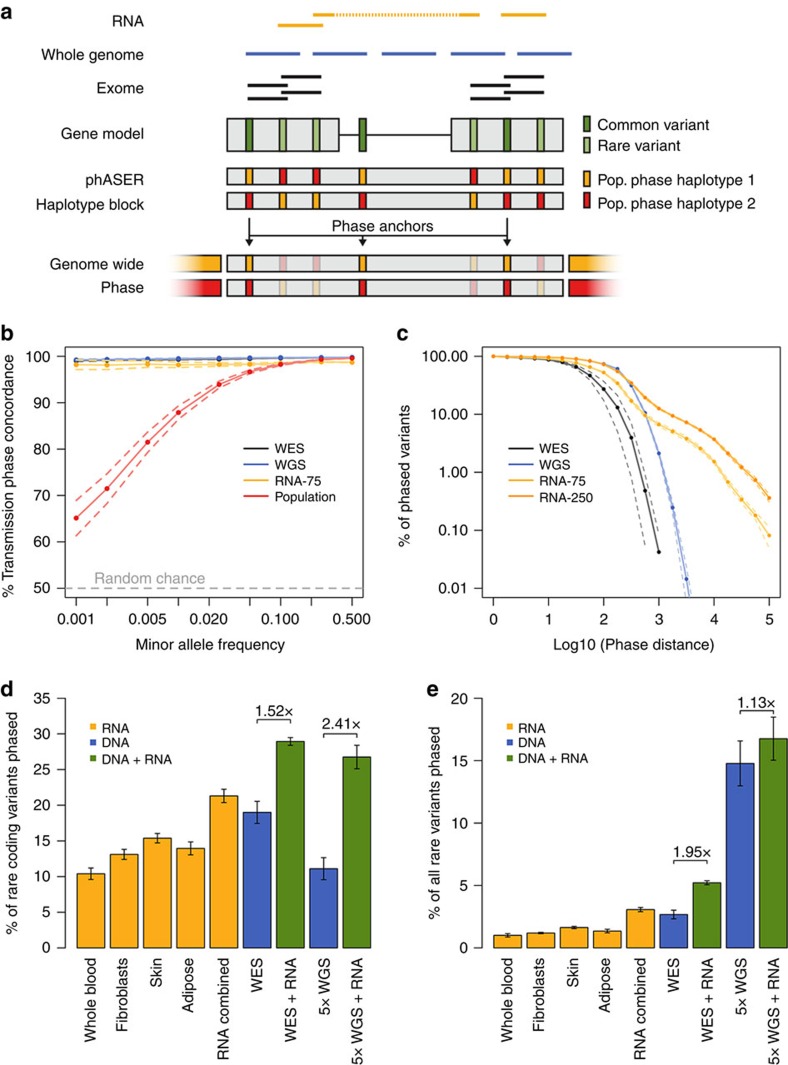
Figure 1. Read backed haplotype phasing that incorporates RNA-seq using phASER.
(a) phASER produces accurate variant phasing through the use of combined DNA and RNA read backed phasing integrated with population phasing. Due to splicing, RNA-seq reads often span exons and UTRs, allowing read backed phasing over long ranges, while high coverage exome and whole genome sequencing can phase close proximity variants. For each group of read connected variants a local haplotype is produced by testing all possible phase configurations, and selecting the configuration with the most support (Supplementary Fig. 1). Local haplotype blocks can be phased relative to one another when population data is available by anchoring the phase to common variants, where the population phase is likely correct. (b) Concordance of read backed phasing across sequencing assays and population phasing with phasing by transmission using the Illumina NA12878 Platinum Genome as a function of variant minor allele frequency. Concordance is defined per variant as the percentage of variant—variant phase events that are correct as compared with the known transmission phase. (c) Percentage of phased variants that can be phased at greater than or equal to increasing genomic distances using WES, WGS, paired-end 75 and 250 RNA-seq data in two tissues (whole blood and LCLs) of four GTEx individuals. Solid lines represent the means, and dotted lines the standard error. (d,e) Contribution of read backed phasing at rare coding (MAF≤1%) variants (d) and all rare variants (e) across sequencing assays and GTEx RNA-seq tissue types for four individuals. Values shown are the mean percentage of rare variants within an individual that can be assigned a genome wide phase using phase anchoring. Error bars show the standard error. The fold increase in the number of rare variants that can be phased using DNA-seq with the addition of combined RNA-seq libraries is indicated.