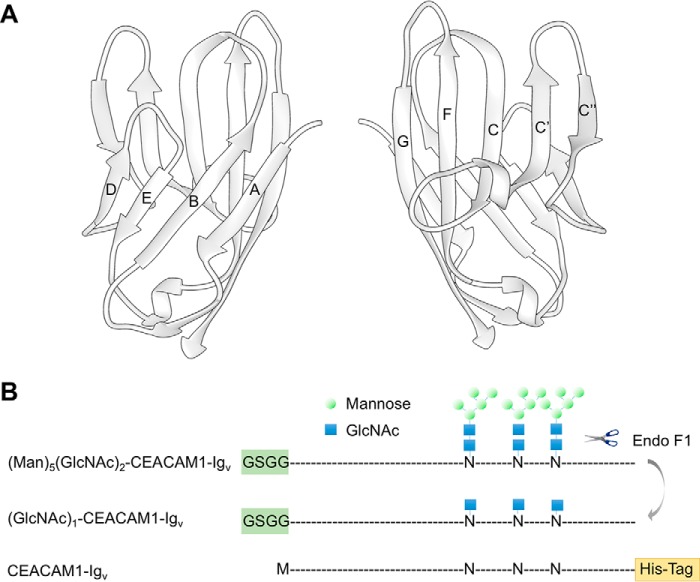
FIGURE 1.
Schematics depicting the structure of hCEACAM1-Igv and the constructs of hCEACAM1-Igv proteins used in this article. A, ribbon diagram of the crystal structure of the CEACAM1 N-terminal Igv domain (PDB code 4QXW). Nine β-sheets are labeled as A, B, C, C′, C″, D, E, F, and G, and they form two distinct faces, which can participate in dimer formation, ABED and GFCC′C″. The former makes dimerization contacts in the 2GK2 crystal structure and the latter makes dimerization contacts in the 4QXW crystal structure. B, CEACAM1-Igv constructs with approximate positions of glycosylation shown as high mannose glycans for the initial expression in HEK293S (GnT1−) cells, as a single GlcNAc after endoglycosidase F1 digestion of this product, and as a non-glycosylated asparagine (N) when produced in E. coli. The blue squares and green circles represent GlcNAc and mannose, respectively. Differences in the constructs at the N and C terminus are also denoted.